Genome-Wide Single-Cell Analysis of Recombination Activity and de novo Mutation Rates in Human Sperm (original) (raw)
. Author manuscript; available in PMC: 2013 Jan 20.
SUMMARY
Meiotic recombination and de novo mutation are the two main contributions towards gamete genome diversity, and many questions remain about how an individual human’s genome is edited by these two processes. Here, we describe a high-throughput method for single-cell whole-genome analysis which was used to measure the genomic diversity in one individual’s gamete genomes. A microfluidic system was used for highly parallel sample processing and to minimize non-specific amplification. High-density genotyping results from 91 single cells were used to create a personal recombination map, which was consistent with population-wide data at low resolution but revealed significant differences from pedigree data at higher resolution. We used the data to test for meiotic drive and found evidence for gene conversion. High throughput sequencing on 31 single cells was used to measure the frequency of large-scale genome instability, and deeper sequencing of eight single cells revealed de novo mutation rates with distinct characteristics.
INTRODUCTION
Gametogenesis is a biological process by which precursor cells undergo cell division and differentiation to form mature haploid gametes. Human gametogenesis occurs by mitotic division of gametogonia, followed by meiotic division of gametocytes into various gametes. During this process the gamete genome experiences both programmed and spontaneous changes, among which meiotic recombination shuffles the two haploid somatic genomes to create a unique hybrid haploid genome for each gamete cell, while accumulated replication errors contribute point mutations which may affect the gametes’ functionality. This results in an enormous variety of new genomes being created in the gametes, thereby enables one’s children to add to the genetic diversity of the human race in a more complex manner than simply mixing and matching entire parental chromosomes. The genome-wide recombination activity and de novo mutation rate have been directly characterized in many model organisms. However, due to ethical and technological challenges, it has been unclear how an individual human’s genome is edited during gametogenesis.
Using pedigree data and statistical methods, deCODE (Kong et al., 2010) and the International HapMap Consortium (The International HapMap, 2005) have been able to create high-resolution recombination maps at the population level. However, such maps only show average results across a population and cumulative results throughout evolutionary history (Jeffreys et al., 2005), and it is not clear what the relationship is between these population maps and the personal recombination processes for any given individual, especially since these focus only on meiotic products that yield successful offspring (Tiemann-Boege et al., 2006). The 1000 Genome Project measured the mutation rate in two family trios (Conrad et al., 2011). However, their results are limited to measuring only a single meiosis per individual, and in general such an approach probes only viable offspring, is limited by the number of offspring per family and requires access to parental genome data.
Here, we describe a single-cell whole-genome analysis method to characterize the genomic changes from gametogenesis. Using this technique, we analyzed the whole genomes of over one hundred single human sperm cells. Recombination data from 91 single sperm cells presented a comprehensive landscape of personal recombination activity. Genome-wide meiotic drive and gene conversion were also directly tested. Single cell whole genome sequencing further revealed primary information about human sperm genome instability and mutation rate.
RESULTS
Microfluidic Single-Sperm Whole-Genome Amplification
We developed a strategy to perform parallel analysis of the haploid genomes of many individual sperm cells by employing single sperm whole genome amplification on a microfluidic device (Figure 1). Previously, we used microfluidic automation to perform whole-genome haplotype analysis by amplifying individual chromosomes at a rate of one cell per device (Fan et al., 2011), and demonstrated high fidelity single chromosome amplification. We have now extended that principle both in parallelization and in complexity of the starting material. The device described here enables the random dispensing of cell aliquots into 48 separate chambers, leading to typically half of them holding exactly one cell. We performed high fidelity amplification of the entire genome in each chamber, followed by whole genome genotyping and high-throughput sequencing analyses.
Figure 1.
Microfluidic device designed for the whole genome amplification from single sperm cells.
Device layout and operation pipeline are slightly modified from a similar device used to measure haplotype. A single sperm cell highlighted by the red square is recognized microscopically and captured in the cross region. In the overview image of the device, control channels are filled with green dye, and flow channels are filled with red dye. See also Table S1.
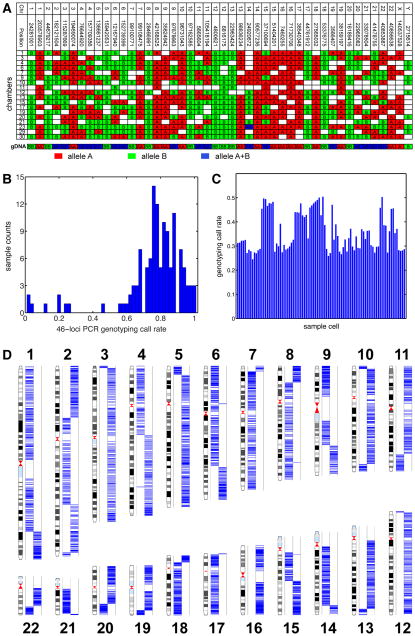
We collected a sperm sample from a 40-year-old Caucasian individual whose genome has been sequenced (Pushkarev et al., 2009), clinically annotated (Ashley et al., 2010) and haplotype phased (Fan et al., 2011) (P0). The patient has healthy offspring and normal clinical semen analysis results. Before the amplification reaction we verified which microfluidic chambers held sperm cells with optical microscopy (Figure 1). With the products of each of the 125 single cell amplification attempts, we performed 46-loci genotyping Taqman PCR to evaluate the amplification performance (a total of 5,750 PCR reactions, a subset of which is shown in Figure 2A). Across the 125 samples, the mean call rate is 76.5% (4398 out of 5750), and 98 samples yielded call rates above 70%, indicating effective whole genome amplification (Figure 2B). 8 samples gave signals in less than 30% of the PCR assays (Figure 2A, chamber 11), suggesting amplification failure or mis-identification of sperm cells by imaging. Because of the haploid nature of sperm cells, amplification products from single sperm cells should give only homozygous genotyping result regardless the polymorphism status of the diploid genome. As expected, 99.4% of the positive PCR reactions yielded signals from only one allele, and the allele combinations from multiple amplification products at each position match the genomic genotype at that locus. The 26 heterozygous calls (0.6% of 4398) reside in 11 of the 125 single cell experiments (ranging from 1 to 7 per cell), and we interpreted these heterozygous calls as the consequence of multiple cells in the chamber or other DNA contamination (Figure 2A, chamber 23). These results show that it is possible to obtain large numbers of high quality single cell genome amplification products using an automated microfluidic device and the products can be used for downstream genomic analysis (Table S1).
Figure 2.
Whole-genome single sperm typing.
(A) Evaluation of amplification performance using 46-loci PCR. This table represents results from a subset of sperm cells being amplified. Each row represents the content from a microfluidic chamber, and each column represents a locus, with specified chromosome number and coordination (NCBI b36). The genotypes of genomic DNA control are also shown. The two alleles of a SNP are highlighted in red and green. Heterozygous loci are labeled in blue. Sample 11 shows a genotyping profile similar to no-template WGA control, indicating mis-identification of sperm cell before amplification. Sample 23 shows heterozygous genotype on chromosome 14 and sex chromosome, suggesting multiple cells during amplification.
(B) 46-loci PCR genotyping call rates.
(C) Whole genome genotyping call rates of 91 single sperm samples from Illumina HumanOmni1S Bead Array.
(D) Detection of recombination from a single sperm sample. The two columns in each chromosome represent the two somatic haplotypes, and blue lines show the genotyping calls of heterozygous SNPs from the sample. Each switch of haplotype block indicates a recombination event.
See also Table S2.
Whole-Genome Sperm Typing from 91 Single Cells Gave a Personal Recombination Map
We selected 93 amplification products with high yield and no heterozygous genotyping calls for an additional round of MDA, followed by Illumina Omni1S whole-genome genotyping (Table S1). Each single cell yielded successful calls at ~30–50% of the 1.2 million SNPs tested (Figure 2C), of which 83.2% were called as homozygous. The lower call rate on the bead array as compared to genotyping PCR is due to amplification bias from MDA. The abundance variation across different regions of the genome exceeds the dynamic range of microarray, and the underrepresented loci are not detected. Taqman PCR, which has much larger dynamic range, gave >70% call rate and this reveals the true extent of coverage of the amplification products. The heterozygous false positive rate is due to similar effects. Within the 0 to ~3 Illumina signal intensity spectrum, the mean intensity of homozygous calls was 1.27 while the mean of heterozygous calls was 0.12, which is barely above the default noise cutoff value of 0.1. These results, together with those from qPCR, reveal that the the heterozygous calls are false positives due to low signal intensity. To improve the genotyping accuracy, we applied a stringent noise cut-off on the raw genotyping calls to remove the low intensity signals and hence eliminate the heterozygous calls.
By mapping the genotyping results from each sperm cell to the two somatic haplotypes obtained by microfluidic direct deterministic phasing (DDP) of single lymphocytes (Fan et al., 2011), we detected single chromosome deletions in two cells (Figure 5A), whereas the other 91 cells gave a total of 2,075 autosomal crossover events (22.8±0.4 SE (±3.7 SD) in each sperm) (Figure 2D and Table S2). The sizes of crossovers range from a few hundred base pairs to over 1 Mbp, with 59%, 37% and 13% of the total events localized to intervals of 200 kb, 100 kb and 30 kb respectively, comparing to 70%, 51% and 20% from previous Hutterite pedigree data for the same intervals. The fact that P0 has a low number of heterozygous loci in the genotyping panel, in combination with the genotyping calling rate, contributed to the slightly lower resolution of our data. The collection of all these recombination events yields a personal recombination map for P0. To our knowledge, this is the first reported high-resolution genome-wide personal recombination map for an individual.
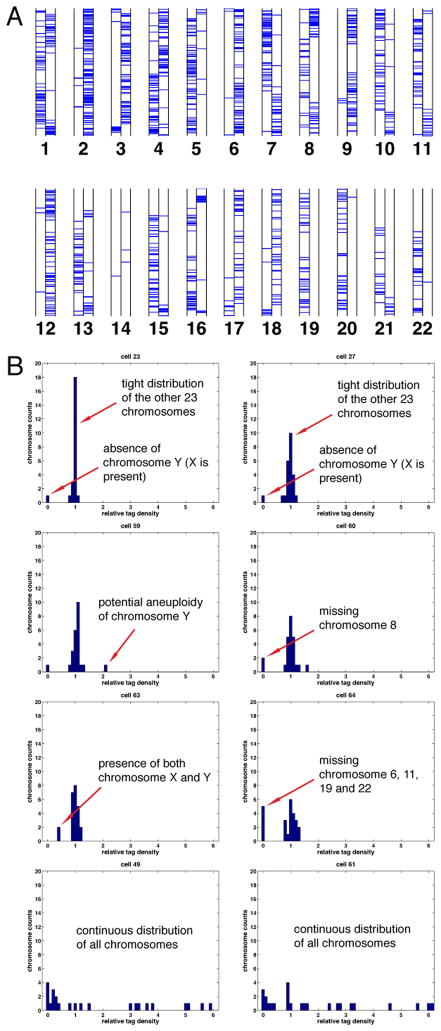
Figure 5.
Germline genome instability.
(A) Whole-genome genotyping results of cell 112. Two columns in each chromosome represent the two haplotypes and each horizontal bar shows the genotype of a SNP. Chromosome 14 showed very low call rates, suggesting its complete deletion.
(B) Cell 23 and 27 are shown as normal controls, with 23 chromosomes clustered by normalized tag density and one sex chromosome dropped. Cells 59, 60, 63 and 64 had whole chromosome aneuploidy. Cell 49 and 61 displayed complex, continuous distributions of chromosome representation.
See also Figure S3.
Personal Recombination Map Recapitulates Population Results at Broad Scale
At a genome-wide scale, the recombination rate of 22.8±0.4 SE (±3.7 SD) events per cell agrees well with the average male results implied from other methods, such as cytological imaging (49.8±0.4 SE (±4.3 SD) MLH1 loci within the tetraploid spermatocytes (Sun et al., 2004)) and data inference (24.0±0.2 SE (±2.7 SD) from Caucasian pedigrees (Cheung et al., 2007)). The slightly lower recombination level in P0 is consistent with his genotype of RNF212 (T/T at rs3796619), which is associated with a 5% lower recombination level than average (Kong et al., 2008). When comparing the number of recombination events within each chromosome, we found similar discrepancies between chromosome length in base pairs and recombination rate (Table S3) as have been previously reported by both cytological and pedigree studies (Sun et al., 2004; The International HapMap, 2005).
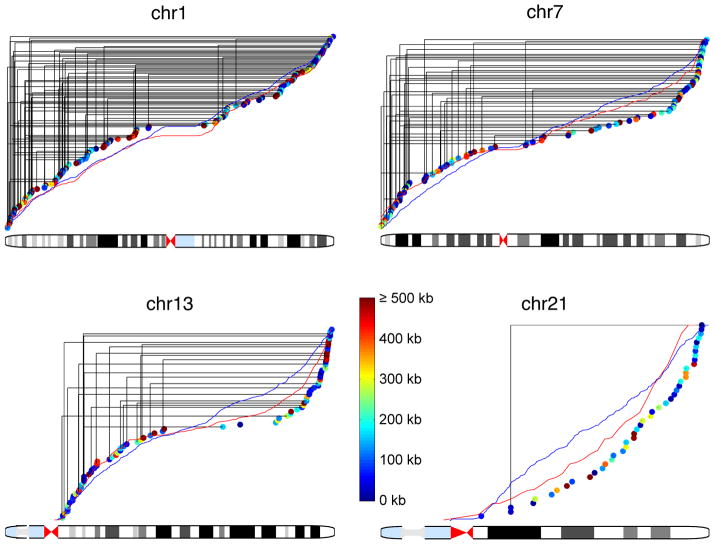
Non-uniformity in the probability of recombination events also occurs within each chromosome. Our data show telomere-weighted distributions that are qualitatively similar to those found in population genetics studies (Kong et al., 2010; Myers et al., 2005). With a 5-Mb window size, we detected a correlation of 0.85 between P0 and deCODE male data, and 0.76 between P0 and HapMap data, while the correlation between deCODE male and HapMap data is 0.85 (Figures 3 and S1). We observed an 87 Mb median distance between adjacent recombination events, comparing with the 49 Mb expected value after we randomly shuffled the recombination events (permutation test, p<10−4), which demonstrates positive recombination interference as has been previously observed (Sun et al., 2004). Taken together, P0’s personal recombination map shows that recombination events within an individual recapitulate the general broad scale features from population data. Our results for the first time experimentally demonstrate general concordance between an individual and the population-average, which can be thought of as an analogy to the ergodic principle from statistical physics.
Figure 3.
Recombination map from chromosome 1, 7, 13 and 21.
Each dot represents a recombination event with color code for resolution. Solid black lines connect recombination events from the same sperm cell. Red and blue lines show the cumulative recombination rates from deCODE (male) and HapMap, respectively. See also Figure S1 and Table S3.
High-Resolution Analysis Revealed Personal Specific Recombination Activity
When one compares our results and the population data at higher resolution, differences emerge. The telomere-weighted bias is stronger in our results than in HapMap or deCODE data, resulting in large regions near the centromere without recombination (Figure S1). For example, no recombination was detected within any ~8 Mb region symmetrically crossing the 17 metacentric chromosome centromeres in P0 (p-value 0.028 based on deCODE male data). The relative activities on the p-arms of some chromosomes are also higher than population-wide results (Figure S1). These differences suggest some potential individual specific features that may be diluted by population-wide averaging, and we therefore performed a more extensive comparison at a finer scale. A sliding window of 2 Mb was applied to P0’s recombination map with 1 Mb increments, and the resulting windows for which P0’s recombination rate was at least triple the genome-wide average (3 cM/Mb) were compared with deCODE male activity. Within the total of 66 such windows, 3 showed significantly higher activity than the deCODE male data in the corresponding regions. We refined the boundaries of these regions and summarized the activities in Table 1 (Sliding Window Scanning).
Table 1.
Examples of Individual Specific Recombination Active Regions in P0
| Chr | Start | End | Size (kb) | P0 Events | deCODE Male (cM)/p-valuea | HapMap Sex-ave (cM)/p-valuea | P0 Activity by Allelic PCR (cM) | P0 Activity by 2-loci Typing (cM) |
|---|---|---|---|---|---|---|---|---|
| 2-Mb sliding window scanning (66 comparisons) | ||||||||
| 20 | 58,753,528 | 59,651,027 | 897 | 12 | 0/<4.7×10−15 | 3.3753/3.8×10−3 | - | - |
| 10 | 20,012,520 | 21,553,899 | 1541 | 6 | 1.175/0.049 | 0.7817/5.7×10−3 | - | - |
| 11 | 132,525,133 | 133,419,195 | 1393 | 14 | 3.97b/9.5×10−4 | 2.3364/1.9×10−6 | - | - |
| P0 self-overlapping sets (324 comparisons) | ||||||||
| 16 | 7,988,699 | 7,990,230 | 1.5 | 2 | 0.12/1.00 | 0.0407/0.21 | 0.77 | 0.84 |
| 9 | 1,864,696 | 1,868,831 | 4.1 | 2 | 0.019b/0.05 | 0.1090/1.00 | - | 0.81 |
| 20 | 58,753,528 | 59,651,027 | 897 | 12 | <1×10−6/<1.5×10−12 | 3.3753/0.019 | - | - |
| 15 | 21,445,700 | 21,732,000 | 286 | 3 | <1×10−6/<1.5×10−12 | 1.2007/1.00 | - | - |
| Non-hotspot overlappingc (135 comparisons) | ||||||||
| 3 | 197,249,108 | 197,250,198 | 1.1 | 1 | 0.0035b/1.00 | 0.0027/0.33 | NDd | - |
| 4 | 18,404,324 | 18,406,601 | 2.3 | 1 | <1×10−6/<1.2×10−4 | 0.0072/0.88 | 1.0 | 1.2 |
Both the deCODE and HapMap projects have made extensive catalogues of recombination hotspots at the population level (Kong et al., 2010; The International HapMap, 2005). Previous sperm studies have demonstrated that some particular hotspots are used idiosyncratically among individuals, but have not had the ability to measure genome-wide activity for an individual (Tiemann-Boege et al., 2006; Webb et al., 2008). Data from a Hutterite pedigree suggested inter-individual variation in hotspot usage (Coop et al., 2008) and supported a hypothesis that the meiosis specific histone methyltransferase PRDM9 may act as a universal regulator for recombination distribution (Baudat et al., 2010). Polymorphisms in PRDM9, to some extent, correlate with the level of historical hotspot usage. However, the small number of meioses each individual has in the pedigree data, as well as uncertainty from statistical haplotype inference, led to extensive overlapping of the hotspot usage percentage between individuals (95% confidence interval of single measurement covering ±25–40%). Consequently, the power of PRDM9 explaining hotspot usage variation is still under debate.
Sanger sequencing showed that P0 has the homozygous A/A PRDM9 genotype (allele naming from (Baudat et al., 2010)), which correlates with the highest historical hotspot usage. We employed the likelihood method from the Hutterite study (Coop et al., 2008) on the portion of P0’s recombination data which matched their criteria (specifically the 274 events with 30kb or smaller size) and determined that only 58% of P0’s recombination events coincide with HapMap hotspots. The 10 times larger sample size in our data led to higher accuracy than the previous results, revealed by our 95% confidence interval of hotspot overlap fraction as ±10%. These high-accuracy measurement of P0’s usage of historical hotspot reveals that even with the most active and hotspot-correlated variant of PRDM9, an individual still generates a substantial proportion of recombination events outside historical hotspots.
We then analyzed the reference human genome for the PRDM9 13-bp degenerate DNA sequence motif which was previously shown to be enriched in HapMap hotspots (Myers et al., 2010; Myers et al., 2008). The motif is significantly (p<10−3) enriched in P0 recombination regions compared to the genome background. However, 50 out of 162 recombination regions smaller than 30 kb do not contain the motif. When we focused on recombination smaller than 10 kb, the enrichment was not significant (p=0.29) due to the low motif occurrence. We performed a de novo motif search within those regions without the 13-bp motif. All five hits reside in transposon sequences, and are significantly enriched in P0’s recombination regions (p<0.05 by simulation). This is consistent with the PRDM9 motif, which is also often located in transposon regions. These results suggest that PRDM9 binding may not be directly required for recombination, and other regulatory mechanisms may exist, such as homologous DNA pairing within transposons.
Among the 2075 recombination events in P0, 940 overlap with at least one another event. These 940 overlapping events form 324 distinct sets, with 2–14 overlapping events in each set. A simulation based on HapMap activities showed significant higher level of self-overlapping in P0 (permutation test, p-value 0.001), suggesting that these recombination clusters are new hotspots. To confirm that P0 does have high recombination activities within these regions, we selected two regions with manageable sizes for allelic PCR and 2-loci digital haplotyping (Figure S2) and independently verified their high activities in P0 (Table 1, Self-Overlapping Sets, chr16:7,988,699–7,990,230, and chr9:1,864,696–1,868,831). By comparing to the deCODE male data, we found that most of these clusters are also active in the population. However, three regions showed significant higher activities than deCODE (Table 1, Self-Overlapping Sets). Considering the small number of recombination events we detected in P0 comparing with the historical hotspots pool, such a high level of overlap demonstrates P0’s preference for only a subset of historical hotspots.
135 of P0’s recombination events do not overlap with any HapMap hotspots. Despite being all singlets, 38 of these events showed statistical significance relative to the activities measured in the deCODE male data, even after multiple comparison adjustment. Such a set as a whole is likely enriched with new recombination spots which can serve as targets for further analysis with traditional sperm typing methods. To demonstrate this, we selected two further regions for allelic specific PCR sperm typing (Figure S2) and discovered that one of them is a new personal hot spot (Table 1, Non-Hotspot Overlapping, chr3:197,249,108–197,250,198 and chr4:18,404,324–18,406,601).
Meiotic Drive and Gene Conversion
Mendel’s laws propose that the two alleles at a genetic position are transmitted to offspring with equal probability. However, results from specific regions and the whole genome have suggested transmission biased toward one allele (Williams et al., 1993; Zöllner et al., 2004), an effect which can in part be explained by the phenomenon of meiotic drive and which can be directly tested in our data.
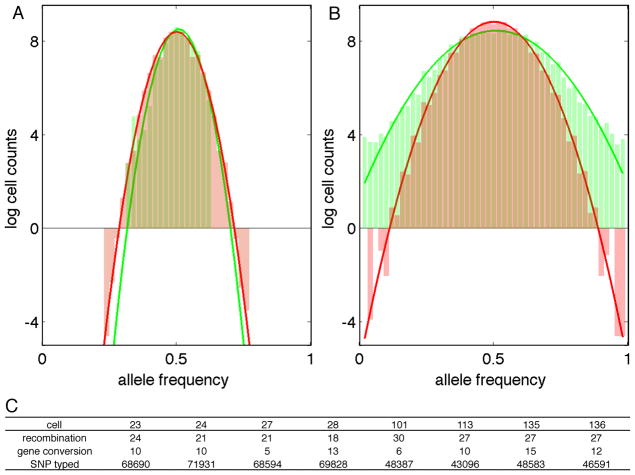
We first investigated if the meiotic drive happens at the whole chromosome level. Because of the general absence of recombination near centromeres, we can accurately define the haplotype across these regions, where kinetochores assemble for mechanical segregation. None of the 22 autosomes presented transmission ratio which significantly deviated from an equal distribution (p>0.7, binomial distribution). Pearson correlation test between different chromosomes didn’t detect any co-transmission of centromere haplotypes. Then we divided the whole genome into 100-kb haplo-blocks and studied if any block showed meiotic drive. Even though many blocks had some evidence for bias, none of them reached genome-wide significance level (Figure 4A). Together with the centromere data, our haplotype block results demonstrate that meiotic drive does not appear as a large haplotype. We then turned to measure the transmission ratio of individual SNPs, where we found obvious difference between our data and simulations of equal transmission (Figure 4B). A putative reason for this pattern is gene conversion.
Figure 4.
Meiotic drive and gene conversion.
Allele frequency histograms of 100-kb haplotype blocks (A) and individual heterozygous SNP (B). Green columns represent experiment data and red columns represent simulation results assuming no transmission distortion. Solid lines are normal distribution fitting results in log scale.
(C) Gene conversion statistics of single cells.
See also Table S4.
Meiotic gene conversion is the transfer of information between homologues without reciprocal recombination. While effectively contributing to genome diversity equally as a pair of closely spaced crossovers, gene conversion is less well studied in human due to its small size relative to genetic marker density. Gene conversion at specific loci has been studied by sperm typing and population genetics data (Gay et al., 2007; Jeffreys and May, 2004), but direct whole-genome measurements have not been conducted for humans.
As shown in Figure 2D, some SNPs have the genotypes the opposite to the haplotype they resided in and therefore serve as good candidates for gene conversion detection. To eliminate potential errors in genotyping, we performed high throughput sequencing on 8 of these cells (Table S1, samples 23, 24, 27, 28, 101, 113, 135 and 136). 6–8× average coverage was obtained with Illumina 2×100 read pairs from each sample, covering ~30–50% of the haploid genome. The less than expected physical coverage based on Poisson statistics is mainly due to amplification bias from MDA. Since the sperm genomes are haploid, one can make highly confident allele calls with substantially lower coverage than the 30× standard genome sequencing depth. To test the accuracy of this genotype calling method, we performed quality control analysis by mapping the sequencing data to the two P0 somatic haplotypes. We correctly detected 184 of the 193 crossover events in these eight cells without false positives, and the nine missing events all reside near the tips of the chromosomes and had low sequencing coverage.
For each gene conversion candidate SNP covered by high-throughput sequencing, we compared the genotypes of the same SNP across different single cells as well as P0 genomic DNA sequencing data (Pushkarev et al., 2009) to confirm genotying and haplotyping accuracy. From the 568 candidates, we confirmed 90 converted SNPs (Table S4). Most gene conversions presented as single SNP, whereas five groups of nearby SNPs gave out gene conversion regions whose sizes from 1 to 22 kbp. This size range is comparable to what was found in yeast (Mancera et al., 2008) but not in human (Jeffreys and May, 2004). More interestingly, when we aligned the converted SNP to historical recombination hotspots, only ten out of the 90 SNPs reside in hotspot regions. This is substantially different from the 58% hotspot overlapping of P0 recombination events. We did not find a strict relationship between gene conversion and recombination level, but generally, cells with more crossovers ended with fewer gene conversions, or vice versa (Figure 4C).
Single-Cell Sequencing Results Reveal Sperm Genome Instability
We chose 8 sperm cells which clearly passed the SNP-PCR assay (“normal”) and 23 further sperm cells which had marginal or failing scores on the assay (“abnormal” and not within the 93 samples for the recombination study) for high throughput sequencing (Table S1) and obtained 0.02× coverage of the genome. After mapping the sequence reads to the human reference genome, we found a discrete distribution of relative sequencing tag density in each chromosome in which chromosomes were typically either present at a uniform level or completely absent (Figure 5B and S3). All the 8 “normal” cells and 17 “abnormal” cells exhibited such patterns with one of the two sex chromosomes missing, while another four “abnormal” cells had clear aneuploidy. Two cells displayed complex, continuous distributions of chromosome representation (Figure 5B and S3). Additional genotyping results confirmed the sequencing findings. The results of these 6 abnormal cells cannot be explained by the known bias mechanisms in MDA (Marcy et al., 2007), and our previous study on single chromosome amplification showed no bias for particular chromosomes or sharp coverage drops in any region (Fan et al., 2011). Therefore the most likely source of missing sequencing reads in the present results is genomic abnormality in the individual sperm cells. The six abnormal samples (Figure 5B), together with the other 2 samples from the recombination analysis (Figure 5A), represent ~7% of the 116 single cell amplifications with high-resolution analysis, which agrees with literature results on aneuploidy of ~2–10% measured with FISH (Luetjens et al., 2002; Macklon et al., 2002).
Human reproduction is well known to be inefficient, with monthly fecundity rates of only 30–40%, and a large number of conceptions fail before the women are aware of the pregnancy (Macklon et al., 2002). This early determination of pregnancy fate was further confirmed by results showing the ability to predict embryo development by the 4-cell stage, before embryonic genome activation (EGA) (Wong et al., 2010). The importance of cytokinesis dynamics in embryo development strongly suggests genome integrity as a key factor, since genome instability will induce cell cycle arrest. Although the aneuploidy rate for oocytes (20%–30%) is higher than that found in sperm (2%–10%), male genome defects are still a significant contribution to conception failure. Even if the embryo does develop correctly, gamete genome abnormality may impose increased risk to certain diseases. For example, the large-scale deletion of chromosome 13 long arm (13q) found in two of our sperm samples (Figure S3B) may induce 13q deletion syndrome with malformations of craniofacial region and skeletal abnormalities (QuÈlin et al., 2009).
de novo Mutations in Primary Sperm Cells
Sequencing data from the gene conversion study also offered the opportunity to measure de novo germline mutations. The recombination detection performed above demonstrated robust genotyping by single cell sequencing and we further evaluated the error rate for mutation detection. We selected high confidence homozygous positions in the P0 somatic genome based on previous sequencing and genotyping (Pushkarev et al., 2009), and calculated the first alternate allele calling frequency in sperm sequencing reads at the same positions. Histogramming these frequency data revealed a decreasing number of positions extending from the perfect agreement side of the discordance axis (Figure S4A). This long tail of background noise represented an amplification/sequencing error rate of 2.7×10−4 per read per position.
A distinct group of loci with 100% discordance with somatic DNA clearly stand out from the amplification error background (Figure S4A). These data are not statistically consistent with any of the measured amplification or sequencing errors and are strong candidates for de novo mutations in the sperm. After excluding signals from repetitive regions or with low alignment confidence, we detected 25 to 36 candidate point mutations in each sperm cell (Table 2 and S5). We selected 19 mutations for PCR-Sanger sequencing and were able to obtain PCR products from 16 regions. The Sanger results from these 16 regions all confirmed our original calls, thus ruling out the possibility of sequence or alignment errors. Since these loci are inconsistent with the statistical distribution of amplification errors, we conclude that they are de novo mutations.
Table 2.
Sperm Mutation Properties
| Sperm ID | 23 | 24 | 27 | 28 | 101 | 113 | 135 | 136 |
|---|---|---|---|---|---|---|---|---|
| Physical coverage (Mbp) | 931 | 987 | 966 | 954 | 1272 | 1231 | 1250 | 1382 |
| Mutation counts | 36 | 30 | 35 | 33 | 27 | 34 | 25 | 31 |
| Mutation rates (×10−8) | 3.8 | 3.0 | 3.6 | 3.5 | 2.1 | 2.8 | 2.0 | 2.2 |
| Ts:Tv | 11.0 | 6.5 | 4.0 | 2.7 | 12.5 | 7.5 | 11.5 | 3.4 |
| CpG | 0.06 | 0.07 | 0.09 | 0.15 | 0.04 | 0.09 | 0 | 0.10 |
| Coding-missense | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| Coding-synonmous | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| UTR | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Non-coding genes | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Intronic | 18 | 7 | 16 | 6 | 13 | 16 | 9 | 11 |
| Intergenic | 17 | 22 | 18 | 26 | 13 | 17 | 16 | 19 |
P0’s mutation rate (2–4×10−8) is higher than that obtained from genome sequenced pedigree data (~1×10−8) (Conrad et al., 2011), but it is consistent with evolutionary studies which have revealed ~4–5x more mutations in male than in female, possibly due to the larger number of germline cell division in male (Crow, 2000; Makova and Li, 2002). The results from the pedigree study identify the variation of germline mutation levels transmitted to each offspring, but are not able to identify the source of such variation. Our results from the eight individual sperm cells have a high degree of internal consistency between their respective mutation levels (Figure S4B), which suggests inter- rather than intra-individual variation. Within each cell, most mutations reside in intergenic or intronic regions (Table 2). However, we detected three missense mutations, a category which was not observed in the pedigree genomes. The transition to transversion ratio of P0 mutations is 5.6, as compared to a population average of 2.1. The main reason of more transition than transversion is generally thought to be deamination of methylated cytosine, primarily at CpG and potentially in other sequence contexts. The higher level of transition we observed is consistent with this as 21% of C->T mutations correlated with CpApG, althougth only 8% were at CpG sites.
DISCUSSION
Despite the advances in personal genomics thus far (Ashley et al., 2010; Levy et al., 2007; Pushkarev et al., 2009; Wheeler et al., 2008), gamete genome variation within individuals, especially fine scale personal recombination activity and germline mutation rates, have been as yet generally inaccessible. Bulk analysis of sperm cells with PCR offers high-resolution and sensitivity (Jeffreys et al., 2005; Webb et al., 2008) and has been used to demonstrate variable usage of historical recombination hotspots, but is limited to investigating focused areas within the genome. Cytological approaches can be used to study recombination-related effects in individuals, but these studies use gamete progenitor cells instead of sperm and have several limitations: (1) the sample collection requires invasive biopsies, (2) the analysis is performed before the completion of meiosis, so it is not clear if all of the synaptonemal complexes proceed to fully recombine and each progenitor cell analyzed by this method predicts an average result from four future sperm cells, and (3) cytological staining does not allow high resolution molecular analysis such as genotyping or sequencing.
There has been increasing interest in performing single cell genome analysis in human cancers, and one can compare the methods and results used in cancer with those used here for human gamete genomes. One group used FACS to sort individual nuclei from human breast tumors (Navin et al., 2011). The genomes from these nuclei were amplified in microliter volumes and lightly sequenced to ~0.2x coverage. This data was sufficient to construct a rough cell lineage map but did not allow calling of individual bases; rather, low-resolution structural variants were used. Another group used mouth-pipetting to isolate individual cells from hematopoietic and kidney tumor (Hou et al., 2012; Xu et al., 2012), whose genomes were then amplified in microliter volumes. Rather than performing whole genome analyses, these samples were then put through exome amplification and sequencing - effectively obtaining 30x coverage of only 1% of the genome. That data was also used to establish lineage relationships between the cells, this time on the basis of point mutations. Their work reveals one of the challenges of performing single cell analyses on diploid genomes - only 57% of the diploid calls were correct. Without the ability to examine a significant proportion of the whole genome, the studies mentioned above had to rely on high mutation rate to distinguish single cells. As a consequence, none of the methods have been applied to samples other than late stage cancers.
In this study, we applied microfluidics to single-cell whole-genome amplification. This technique not only presents great parallelization, but also improved amplification performance. MDA is sensitive to environmental contamination and extensive sample purification is required for traditional bench-top whole-genome amplifications (Hou et al., 2012; Jiang et al., 2011; Xu et al., 2012). More sensitive assays even revealed contamination in the MDA reagents (Blainey and Quake, 2010). By incorporating the amplification into microfluidic chips, we reduced the reaction volume, and hence the contamination, by ~1000 fold.
Amplification error has been a concern for single cell whole-genome analysis. Previous microliter volume single cell exome studies have shown 2–3×10−5 false discovery rates from MDA (Hou et al., 2012; Xu et al., 2012). Using our microfluidic approach on haploid cells, we have reduced the error rate to 4×10−9 with 5× coverage (binomial probability with per read error rate). An important feature of single molecule MDA is its repetitive usage of the originating genuine template molecule. Even if an amplification error happens in the initial stage, there will still be a large fraction of products preserving the correct base information from the original template, and the power of statistics from multiple coverage discriminates these errors from true genomic variation.
Using this microfluidic MDA approach, we reported the first genome-wide single cell analysis of human sperm. We were able to create a personal recombination map for an individual and to measure the rate of de novo mutations in this individual’s germline. The advantage of sampling a large set of meioses from single individual for fine scale analysis allowed us to uncover individual specific features potentially buried under population data. P0’s preference for a subset of historical hotspots suggests how individual features contribute to the population diversity, and a potential solution for the hotspot paradox. We propose that this partially overlapping feature is also the general pattern in individuals: everyone is using a different subset of the historical hotspots; while some hotspots are dying in some people, new recombination activities evolve to refill the hotspot pool; the partially overlapping patterns of individuals give rise to the population results, with hotspots (still active in many people) and deserts (used by fewer people). Support to this theory comes from single cell analysis. While P0 has on average 58% overlap with the historical hotspots, this ratio range from 0 to 100% for his single cells (Figure S2D). The partially overlapping patterns between individual cells produce P0’s personal recombination landscape.
Transmission distortion has long been known but the key factors behind it are not clear. Biased segregation during meiosis, different ability to achieve fertilization and different postzygotic viability can all contribute to this phenomenon. Specifically if meiotic drive exists, the molecular mechanism is not known. Our data from 91 cells showed that meiotic drive does not generally appear as whole haplotype blocks, but may occur at individual SNP loci. The most intuitive explanation for this result would be gene conversion. Indeed, we found 5–15 gene conversions in each genome sequenced cell. This represents a lower bound for the total number of conversions in each single human sperm, since there is a limited heterozygous SNP density. If both crossover events and gene conversion originate from double strand breaks and share a recombination mechanism, then they should have the same hotspot overlapping ratio. If we match the number of gene conversion at hotspots and further assume there are 1.5 million heterozygous SNP in the genome, the total number of gene conversion in a single cell would be ~250–800, which is 10–35× of crossovers. Previous sperm typing studies have suggested 4–15× gene conversions over crossovers, based on data from 3 hotspots (Jeffreys and May, 2004), but this value apparently changes across the genome (Gay et al., 2007).
Evolutionary studies have estimated the germline mutation level (Makova and Li, 2002) but recent results from 1000 Genome Project (Conrad et al., 2011) are not consistent with the previous findings. The combination of data from our study and the 1000 Genome Project suggest that the germline mutation rate can vary greatly among different individuals but not different cells from the same individual. This may explain why the male mutation rate is not always higher than the female. DNA methylation also affects genome instability (Li et al., 2012) and C->T point mutation levels, but in opposite ways. A fine tuned methylation level is therefore required for high quality sperm genome. The high germline mutation rate at CpA regions (Conrad et al., 2011; Miyoshi et al., 1992) at least suggests a methylation profile different from the somatic genome. The fact that cytosine deamination is less well repaired at CpA than at CpG also explains our findings (Wang and Edelmann, 2006).
The ability to study a large numbers of single sperm cells has offered several new insights in meiosis biology. Studying the germline genome is but one application of single cell genomics and we expect that the method described here will find applications in many other fields, including cancer, aging, immunology and developmental biology.
EXPERIMENTAL PROCEDURES
Sample Collection
Semen sample was collected from P0 in Stanford Reproductive Endocrinology and Infertility Center, and analyzed with a computer assisted semen analyzer following clinical standards.
Single Cell Whole Genome Amplification
Whole genomes from P0’s single sperm cells were amplified on a microfluidic device using multiple strand displacement amplification (Repli-g midi, QIAGEN), yielding a gain of ~104 fold. Amplification products from single sperm cells were subjected to 46-loci Taqman genotyping PCR assays (Applied Biosystems) to evaluate the amplification performance.
Public Data Set Access
Human reference genome sequence and annotation were downloaded from UCSC Genome Bioinformatics (http://genome.ucsc.edu/). The P0 somatic genome and genotyping data were from previous study. Population recombination data were from HapMap Project (http://hapmap.ncbi.nlm.nih.gov/, rel22) and deCODE genetics (http://www.decode.com/).
Personal Recombination Map
93 samples with high amplification efficiency were re-amplified by MDA and genotyped on Illumina’s Omni1S Bead Array. Raw genotyping data were processed by Illumina GenomeStudio for genotype calling and further filtered to remove low intensity calls. Haploid genome from each sperm cell was aligned to the two P0 somatic haplotypes (Fan et al., 2011). Recombination events were called by a MATLAB script and further manually confirmed. Distribution of P0 recombination events along the genome was compared with population-wide data from deCODE (male non-carrier) and the HapMap Project. Statistical analysis with population data was based on binomial distribution, followed by Bonferroni correction. Recombination frequency in selected regions with recurrent crossover events was measured using allelic specific digital PCR (BioMark, Fluidigm).
Meiotic Drive Measurement
Centromere boundaries were extracted from UCSC chromosome band table and were extended by 5Mbp to include enough heterozygous SNP. Chromosome haplotype was defined as the haplotype of its centromere. Allele frequency of each autosome was measured based on recombination data of 91 single cells. Pearson correlation coefficient was calculated for each pair of chromosomes to detect potential co-transmission of chromosome haplotypes. The whole genome was further divided in 100kbp blocks and the haplotype of each block was assigned based on chromosome-wide recombination results. Blocks overlapping with recombination events were excluded to avoid haplotype ambiguity within the recombination region. Binomial distribution was used for no distortion simulation. We studied the allele frequency of single SNP with the same approach.
Gene Conversion
For each of the 8 single cells, SNPs with allele pointing to the other haplotype of the block they reside in were extracted for further examination. The same 8 cells were sequenced on Illumina HiSeq2000 with 2×100 paired-end read option. Raw sequencing data were processed by Illumina software and aligned to human reference genome hg18 with BWA. Alignment was refined with GATK realignment tool and piled up with samtools. For each gene conversion candidate SNP covered by high-throughput sequencing, we set three requirements for gene conversion calling. The SNP must have sequencing support for its allele call; the SNP must have sequencing support from other single cells or P0 genomic DNA for its heterozygosity; the SNP must have support from other single cell for its haplotyping. SNP failed to meet any of the above three requirements would be cataloged as low coverage or genotyping error, low heterozygosity and haplotyping error.
Single Cell Genome Instability Measurement
Single sperm samples with bulk sperm genomic DNA were subjected to multiplex Illumina library construction with NuGEN Encore NGS kit and briefly shotgun sequenced on an Illumina GAII with 1×36 read option. Raw sequencing data were processed by Illumina software and aligned to human reference genome hg18 with CASAVA 1.6.0. Sequencing tag density in every 500-kb non-overlapping window was counted and normalized with sperm genomics DNA control. Genome abnormality was analyzed based on sequencing tag density distribution.
de novo Mutation Detection
To eliminate random errors induced by MDA and sequencing, we used a binomial test to detect high-confidence mutations. Specifically, false positive rate from MDA and sequencing was measured by using high-confidence homozygous loci in P0 from genotyping and somatic sequencing. The probability of observing given number of mutation calls under given sequencing depth was calculated for each position with a Binomial Distribution.
A complete description of the materials and methods is provided in Supplemental Experimental Procedures.
Supplementary Material
01
02
03
04
05
06
HIGHLIGHTS.
- A microfluidic system for high-throughput single cell whole-genome amplification
- Personal recombination map by single sperm whole-genome analysis
- Genome-wide meiotic drive and gene conversion tests
- de novo germline mutations with distinct molecular characteristics
Acknowledgments
We would like to thank Jessica Melin and the Stanford Microfluidic Foundry for fabrication of the microfluidic devices. We thank Janice Gebhardt and the Stanford IVF Laboratory for semen sample processing. We thank Norma Neff, Gary Mantalas, and Ben Passarelli for assistance in sequencing experiments and data processing. We also thank Jad Kanbar for help with PCR experiments. The project was supported by CCNE-T, a grant funded by NCI-NIH (grant no: U54CA151459). J.W. was supported by a scholarship from the Chinese Scholarship Council. H.C.F was supported by a scholarship from the Siebel Foundation.
Footnotes
All sequencing data from this study are deposited in NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the accession number SRA053375.
AUTHOR CONTRIBUTIONS
J.W., H.C.F. and S.R.Q. designed research; B.B. coordinated sperm sample collection; H.C.F. designed microfluidic device; J.W. and H.C.F. performed research; J.W., H.C.F. and S.R.Q. analyzed data and wrote the paper; and all authors discussed results and commented the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, et al. Clinical assessment incorporating a personal genome. The Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 Is a Major Determinant of Meiotic Recombination Hotspots in Humans and Mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Burdick JT, Hirschmann D, Morley M. Polymorphic Variation in Human Meiotic Recombination. American journal of human genetics. 2007;80:526–530. doi: 10.1086/512131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JEM, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-Resolution Mapping of Crossovers Reveals Extensive Variation in Fine-Scale Recombination Patterns Among Humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotech. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay J, Myers S, McVean G. Estimating Meiotic Gene Conversion Rates From Population Genetic Data. Genetics. 2007;177:881–894. doi: 10.1534/genetics.107.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-Cell Exome Sequencing and Monoclonal Evolution of a JAK2-Negative Myeloproliferative Neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat Genet. 2005;37:601–606. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- Jiang N, Weinstein JA, Penland L, White RA, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proceedings of the National Academy of Sciences; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, Jonsdottir GM, Gudjonsson SA, Sverrisson S, Thorlacius T, et al. Sequence Variants in the RNF212 Gene Associate with Genome-Wide Recombination Rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, et al. Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome. PLoS Genet. 2012;8:e1002692. doi: 10.1371/journal.pgen.1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetjens CM, Rolf C, Gassner P, Werny JE, Nieschlag E. Sperm aneuploidy rates in younger and older men. Human Reproduction. 2002;17:1826–1832. doi: 10.1093/humrep/17.7.1826. [DOI] [PubMed] [Google Scholar]
- Macklon NS, Geraedts JPM, Fauser BCJM. Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Human Reproduction Update. 2002;8:333–343. doi: 10.1093/humupd/8.4.333. [DOI] [PubMed] [Google Scholar]
- Makova KD, Li WH. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, Beeson KY, Goldberg SMD, Quake SR. Nanoliter Reactors Improve Multiple Displacement Amplification of Genomes from Single Cells. PLoS Genet. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y, Ando H, Nagase H, Nishisho I, Horii A, Miki Y, Mori T, Utsunomiya J, Baba S, Petersen G. Germ-line mutations of the APC gene in 53 familial adenomatous polyposis patients. Proceedings of the National Academy of Sciences. 1992;89:4452–4456. doi: 10.1073/pnas.89.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A Fine-Scale Map of Recombination Rates and Hotspots Across the Human Genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive Against Hotspot Motifs in Primates Implicates the PRDM9 Gene in Meiotic Recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotech. 2009;27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QuÈlin C, Bendavid C, Dubourg C, de la Rochebrochard C, Lucas J, Henry C, Jaillard S, Loget P, Loeuillet L, Lacombe D, et al. Twelve new patients with 13q deletion syndrome: Genotype-phenotype analyses in progress. European Journal of Medical Genetics. 2009;52:41–46. doi: 10.1016/j.ejmg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Sun F, Oliver-Bonet M, Liehr T, Starke H, Ko E, Rademaker A, Navarro J, Benet J, Martin RH. Human Male Recombination Maps for Individual Chromosomes. American journal of human genetics. 2004;74:521–531. doi: 10.1086/382138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann-Boege I, Calabrese P, Cochran DM, Sokol R, Arnheim N. High-Resolution Recombination Patterns in a Region of Human Chromosome 21 Measured by Sperm Typing. PLoS Genet. 2006;2:e70. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JYJ, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer cell. 2006;9:417–418. doi: 10.1016/j.ccr.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Webb AJ, Berg IL, Jeffreys A. Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proceedings of the National Academy of Sciences. 2008;105:10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Williams C, Davies D, Williamson R. Segregation of δF508 and normal CFTR alleles in human sperm. Human Molecular Genetics. 1993;2:445–448. doi: 10.1093/hmg/2.4.445. [DOI] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, Pera RAR. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotech. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-Cell Exome Sequencing Reveals Single-Nucleotide Mutation Characteristics of a Kidney Tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner S, Wen X, Hanchard NA, Herbert MA, Ober C, Pritchard JK. Evidence for Extensive Transmission Distortion in the Human Genome. American journal of human genetics. 2004;74:62–72. doi: 10.1086/381131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06