Rescue of hearing and vestibular function in a mouse model of human deafness (original) (raw)
. Author manuscript; available in PMC: 2013 Sep 1.
Published in final edited form as: Nat Med. 2013 Feb 4;19(3):345–350. doi: 10.1038/nm.3106
Abstract
Hearing impairment is the most common sensory disorder, with congenital hearing impairment present in ~1 in 1000 newborns1, and yet there is no cellular cure for deafness. Hereditary deafness is often mediated by the developmental failure or degeneration of cochlear hair cells2. Until now, it was not known whether such congenital failures could be mitigated by therapeutic intervention3-5. Here we show that hearing and vestibular function can be rescued in a mouse model of human hereditary deafness. An antisense oligonucleotide (ASO) was used to correct defective pre–mRNA splicing of transcripts from the mutated_USH1C_.216G>A gene, which causes human Usher syndrome (Usher), the leading genetic cause of combined deafness and blindness6,7. Treatment of neonatal mice with a single systemic dose of ASO partially corrects USH1C.216G>A splicing, increases protein expression, improves stereocilia organization in the cochlea, and rescues cochlear hair cells, vestibular function and hearing in mice. Our results demonstrate the therapeutic potential of ASOs in the treatment of deafness and provide evidence that congenital deafness can be effectively overcome by treatment early in development to correct gene expression.
Usher is characterized by hearing impairment combined with retinitis pigmentosa (RP) and, in some cases, vestibular dysfunction. The frequency in the general population, may be as high as 1 in 60008. Type 1 Usher (Usher 1) is characterized by profound hearing impairment and vestibular dysfunction at birth and the development of RP in early adolescence. Approximately 6–8% of Usher 1 cases are caused by mutations in the_USH1C_ gene9, which encodes the protein Harmonin. The USH1C.216G>A (216A) mutation accounts for all cases of Usher 1 in Acadian populations9-11. The 216A mutation creates a cryptic 5′ splice site that is used preferentially over the authentic 5′ splice site of exon 3 (Fig. 1a), resulting in a frameshift and truncated harmonin protein12.
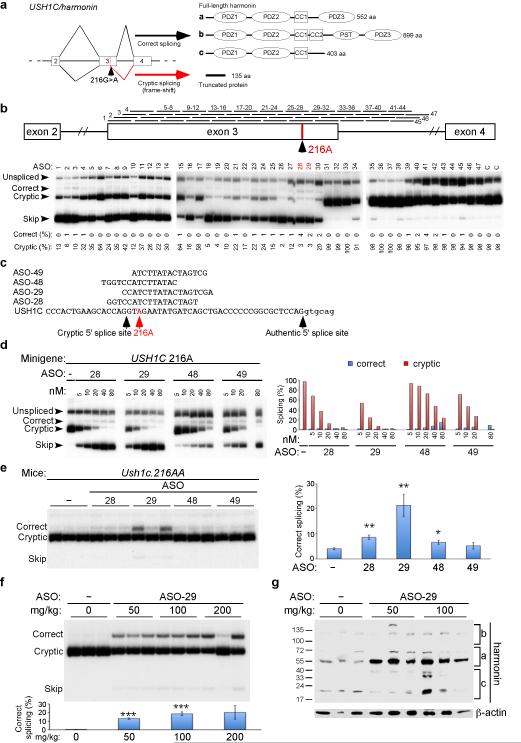
Figure 1. Correction of USH1C.216A splicing using ASOs.
a, USH1C exons 2–4 gene structure, RNA splicing and protein products. Boxes represent exons and lines are introns. Diagonal lines indicate splicing. The locations of the 216A mutation and the cryptic splice site are labeled. b, (top) Diagram of ASOs used in walk, mapped to their position of complementarity on USH1C. (Bottom) Radioactive RT–PCR of RNA isolated from HeLa cells transfected with_USH1C_.216A minigene and indicated ASO at a final concentration of 50 nM. RNA spliced forms are labeled. Unspliced refers to transcripts with intron 3 retained and skip indicates exon 3 skipping. Quantitation of % correct splicing in graph is calculated as [(correct/(correct + cryptic +skip)]*100 and similarly for % cryptic. c, Sequence and_USH1C_ target region of ASOs. d, Analysis of _USH1C216A_minigene transcript splicing in HeLa cells treated with different concentrations of indicated ASOs. Quantitation of % correct and % cryptic splicing is shown in graph (right). **e,**RT–PCR analysis of RNA isolated from kidneys of adult Ush1c 216AA mice injected with 50 mg kg−1 of different ASOs. Samples from three individual mice are shown. Ush1c spliced products are indicated and quantitated in graph (right) as described above. Error bars represent SEM (*P ≦0.05, **_P_≦0.01, n=3, two– tailed Student’s t–test compared to vehicle treatment).f, RT–PCR analysis of RNA isolated from kidneys of adult Ush1c 216AA mice treated with different doses of ASO– 29. Samples from three individual mice are shown. Ush1c spliced products are indicated and quantitated as described above. Error bars represent SEM (***P ≦ 0.001, n=3, two– tailed Student’s t–test compared to vehicle). g, Western blot analysis of harmonin protein in lysates from the kidneys of adult Ush1c 216AA mice analyzed in f. Blots were also probed with a β–actin–specific antibody for a loading reference. ASO, antisense oligonucleotide; nM, nanomolar.
We used a mouse model of Usher based on the human 216A mutation13 to investigate a treatment for deafness and vestibular dysfunction using antisense oligonucleotides (ASO)(Supplementary Fig. 1) designed to redirect cryptic splicing of 216A RNA to the authentic site (Fig. 1a). To screen for ASOs that block 216A cryptic splicing, a minigene expression plasmid comprised of exons 2–4 and the intervening introns of human_USH1C.216G_ (WT) or 216A was transfected into cells with 47 different individual ASOs surrounding the mutation and splicing correction was quantitated (Fig. 1b; Supplementary Table 1). Several ASOs blocked cryptic splicing and promoted correct splicing (Fig. 1b,c) in a dose-dependent manner (Fig. 1d). The ASOs also blocked cryptic splicing, promoted correct splicing of the endogenous Ush1c.216A gene transcript, and increased harmonin protein expression in a mouse kidney cell line derived from mice homozygous for the Ush1c.216A mutation (Supplementary Fig. 2). ASOs induced correct splicing in vivo following intraperitoneal injections of 50 mg kg−1 of ASO in adult Ush1c.216AA mice. ASO–29 promoted the highest amount of correct splicing of the ASOs tested (Fig. 1e) and also corrected splicing and increased Harmonin protein expression (Fig. 1f,g respectively) in a dose-dependent manner.
Mice homozygous for the Ush1c.216A mutation (216AA), exhibit head–tossing, and circling behavior indicative of vestibular dysfunction and deafness13,14. We gave single intraperitoneal injections of various ASOs to neonatal 216AA mutant mice to test whether ASOs can correct vestibular and hearing defects. 216AA mice untreated or treated with a mismatched ASO (ASO–C) displayed circling behavior whereas mutant mice treated with ASO–29 did not circle, similar to heterozygous (216GA), or wildtype (216GG) mice (Fig. 2a,b, Supplementary Video 1). No circling was observed in mice treated at P3, P5, P10, or P13, whereas P16–treated mutant mice exhibited circling behavior similar to untreated or ASO–C–treated mutant mice (Fig. 2b). ASO-29-treated mutant mice have no vestibular dysfunction at more than 12 months of age (Supplementary Fig. 3a). We performed trunk–curl, contact–righting, and swim tests on 2–3 month and 6–9 month old mice to further quantitate vestibular function15. The younger and older mutant mice all performed poorly in these tests, whereas the mutant mice treated with ASO–29 at P5 performed at the same level as heterozygous mice and showed no vestibular dysfunction (Supplementary Fig. 3b). Our results suggest that ASOs can effectively cure vestibular dysfunction associated with Usher in mice when delivered neonatally.
Figure 2. ASOs correct vestibular function and rescue hearing in Ush1c.216AA mice.
a, Representative open–field pathway trace (120 s) from a P22 mouse in each group are shown. b, Quantitation of the number of rotations in 120 sec. Error bars represent SEM, the number (n) of animals analyzed is indicated within the individualbars. Significance (***P ≦ 0.001 or not significant, NS) was calculated using one–way ANOVA and Tukey–Kramer post test (Graphpad Software, La Jolla, CA). c, Representative ABR waveforms at 8 kHz stimulus from a 216AA mouse injected at P5 with control ASO (ASO–C, left panel), 216AA mouse injected at P5 or P10 with ASO– 29 (middle panels) and a heterozygote 216GA mouse (Heterozygote (GA) Ctl, right panel). Colored lines (red = AA, ASO–C; blue = AA, ASO–29 injected at P5; orange= AA, ASO–29 injected at P10; gray = GA Ctl) represent thresholds detected. Average ABR thresholds (dB SPL) to pure tones ranging from 8 to 32 kHz or BBN in Ush1c.216AA mutant and wildtype (GG) or heterozygous (GA) mice at d, one month, (n =11, 8, 5, and 11 for 216AA, ASO–C; 216AA, ASO–29 at P5; 216AA, ASO–29 at P10 and 216GG/GA, respectively) e, two months, (n = 4, 6, and 5 for 216AA, ASO–C; 216AA, ASO–29; and 216GG/GA, respectively) f, and three months of age (n = 3, 4, and 4 for 216AA, ASO–C; 216AA, ASO–29; and 216GG/GA, respectively). Error bars represent SEM. Asterisks (*) indicate a significant difference between 216AA, ASO–29 treated and 216AA, ASO–C treated mice and a pound sign (#) indicates a significant difference between 216AA, ASO–29 and 216GA control mice (_P_≦ 0.05; two–way ANOVA with Tukey–Kramer post test). dB, decibels; SPL, sound pressure level; kHz, kilohertz; μV, microvolts; msec, milliseconds, BBN, broad band noise.
Treatment of 216AA mice with ASO–29 also rescued hearing. Startle responses to high amplitude sound are similar in ASO–29–treated 216AA and 216GA control mice (Supplementary Video 2; Supplementary Fig. 4). 216AA mice treated with ASO–C, however, exhibited neither an initial startle response, defined as an ear–twitch and rapid head and body movement, nor a subsequent freezing response after acoustic stimulus (Supplementary Video 2). We performed auditory–evoked brainstem response (ABR) analysis to quantitatively assess hearing function. Responses to different sound frequencies (8–32 kHz and broad band noise, BBN) at different intensities (18–90 dB) were recorded. Hearing thresholds represent the lowest sound intensity at which a recognizable ABR wave (neural response) is observed. We compared ABR thresholds in P30 216AA mice treated with ASO–29, treated and untreated wild type and heterozygous mice, and mutants treated with ASO–C. Wild type and heterozygous mice had thresholds typical of mice with normal hearing (Fig. 2c,d). Similar to untreated mutants14, mutants treated with ASO–C had abnormal or absent ABRs (Fig. 2c,d; Supplementary Fig. 5). Mutant mice treated between P3–5 with a single dose of ASO–29 had recognizeable waveforms and near normal thresholds to BBN at 8 and 16 kHz pure tones when compared to wild type and heterozygous control mice (Fig. 2c,d; Supplementary Fig. 5). The thresholds of ASO–29–treated mutants at 32 kHz were not significantly different than control mutants, indicating that treatment was not effective at rescuing high frequency hearing (Fig. 3d). These data show rescue of low and mid frequency hearing at P30. Mutant mice treated with ASO–29 at P10 had significantly higher thresholds than those treated at P3–5, but significantly lower thresholds in response to BBN and an 8 kHz tone than untreated mutants or mutants treated with ASO–C (P ≤ 0.05) (Fig. 2c,d;Supplementary Fig. 5), implicating a developmental window of therapeutic efficacy in mice. At two months of age, mutant mice treated at P3-5 had ABR thresholds to BBN, 8 and 16 kHz, but not 32 kHz, equivalent to wild type and heterozygous mice (P ≤ 0.05, Fig. 2e,Supplementary Fig. 6a–d). At 3 months, there was no significant difference in ABR thresholds at 8 kHz between control heterozygote mice and mutant mice treated with ASO–29 at P3-5 (P ≤ 0.05), but data show some loss of sensitivity to 16 kHz and BBN (Fig. 2f; Supplementary Fig. 7a–d). At 6 months of age, there were significant differences in ABR thresholds between control heterozygote and ASO–29 treated mutant mice at all frequencies (P ≤ 0.05), though 216AA treated mice exhibited ABR thresholds that were significantly lower than ASO–C treated mutants (P ≤ 0.05) (Supplementary Fig. 8a,b). These results show that mice injected with a single ASO treatment early in lifecan hear at 6 months of age, indicating a long–term, if slowly declining, therapeutic correction of deafness.
Figure 3. ASO–29 treatment partially rescues splicing, harmonin protein abundance and cochlear hair cells.
a, RT–PCR analysis of cochlea RNA isolated at P32–P35 from mice treated with control (ASO-C) or ASO–29 at P3–5. Spliced products are labeled.b, Western blot analysis of harmonin in cochlea isolated at P32–35 from mice that were treated at P5. Different isoforms of harmonin expressed from USH1C are indicated. Blots were also probed with a β–actin–specific antibody for a loading reference. c, Immunofluorescence images of Harmonin b (green) and F–actin (red, phalloidin) in OHC bundles in the region of the basilar membrane that corresponds to hearing at 8 kHz (0.8– 1.5 mm from apex tip). Images are from heterozygote mice (left panel), 216AA mice treated with ASO–29 at P5 (middle), or 216AA mice treated with ASO–C at P5 (right). Scale bar=3 microns. d, Images are higher magnification (X 100 original magnification) of the images shown in c. e, Immunofluorescence image of a primary antibody isotype control from a heterozygote mouse taken from a similar OHC bundle location.f, Immunofluorescence image of the regions of the basilar membrane that are represented in c, e, and f. Hair cells are labeled with parvalbumin (red) and nuclei counterstained with DAPI (blue). Scale bar=50 microns. g, Cochleogram showing inner (top) and outer (bottom) hair cells counts from regions progressively distant from the apex tip. Error bars represent the standard error of the least square means. (*P ≤ 0.05; n=3 mice). At least 100 cells from each experimental group were evaluated for each region. h, Immunofluorescence images of representative regions along the basilar membrane 0.8– 1.0 mm (top) or 1.8–2.0 mm (bottom) from the extreme apex from P35 heterozygote (left panels), or 216AA mice treated with ASO–29 at P5 (middle), or 216AA mice treated with ASO–C at P5 (right). OHCs, outer hair cells; IHCs, inner hair cells, mm=millimeter, μm=micron, kHz = kilohertz. Scale bar = 20 microns.
To determine the effect of ASOs on Ush1c RNA splicing and harmonin expression, cochleae from mice injected at P5 with ASO–29 or ASO–C were analyzed. At P30, a low amount of correct exon 3 splicing was observed in the ASO–29–treated 216AA mice (Fig. 3a). Correct splicing peaked at P120 and correction at P180 was similar to that at P30, indicating that the effect of the ASO on splicing is stable and correlates with ABR results (Supplementary Fig. 8c). Harmonin protein abundance also was elevated in cochleae from ASO–29–treated mutant mice compared to mutant mice treated with ASO–C, and similar to harmonin levels in the cochleae of control 216GA mice (Fig. 3b).
The Harmonin b isoform (Fig. 1a) localizes to the developing and mature stereocilia bundle of cochlear hair cells16-19 where it is hypothesized to scaffold the molecular components of the mechanotransduction machinery20. We examined expression and localization of Harmonin b in microdissected Organs of Corti from P30, 216AA mice injected at P5 with either ASO–29 or ASO–C and heterozygote littermates. Harmonin b was abundantly expressed in the outer hair cell stereocilia bundles of heterozygote mice whereas the mutant mice had decreased expression in atypical bundles (Fig. 3c,d). Mutant, ASO-29-treated mice had elevated harmonin b expression with localization at the tips of the stereocilium similar to the heterozygote mice (Fig. 3c,d). These results indicate a molecular correlation to the therapeutic effect of treatment with ASO–29.
Frequency place–mapping in the mouse cochlea21,22 suggests that the region corresponding to 8–16 kHz (the frequencies most robustly rescued by ASO–29) is located approximately 1–2 mm from the apex tip (Fig. 3f, 4b). To assess the relationship between hair cell number and ABR threshold, hair cells were labeled with parvalbumin. At P35, mutant mice had significant outer hair cell loss from approximately 0.8–2.0 mm from the apex, corresponding to hearing at 6–20 kHz (P ≤ 0.05) (Fig. 3g,h). In contrast, the number of outer hair cells in this region of mutant mice treated with ASO–29 at P3-P5 did not differ from heterozygotes, consistent with rescued physiological function (Fig. 3g,h).
Figure 4. Restoration of hair cell bundle morphology in mice.
a, Scanning electron micrographs of outer hair cell bundles from P18 216GA, and 216AA mice treated at P5 with ASO–C or ASO–29. Distance (mm) was measured from apex tip. Scale bars represent 1 μm and 10 μm for the high and low magnification images, respectively. b, Scanning electron micrograph illustrating the regions of the cochlea that are represented in a. c, Quantitation of atypical bundles shown as a percent of total cells counted at different positions along the basilar membrane in mutant mice treated either with ASO– 29–treated mice (blue line) or ASO–C (red line) (*_P_≤ 0.05, **P ≤ 0.005; two–tailed unpaired t–test; n=3–4 mice per region). Error bars represent SEM. At least 200 cells from each experimental group were evaluated with at least 60 hair cells from each region. 216GA control mice have no atypical bundles (data not shown). mm = millimeter, μm = micron, kHz = kilohertz.
We also assessed changes in hair cell morphology that may reflect the rescue of hearing in the regions of the cochlea sensitive to 8 and 16 kHz. By P35, mutant mice have significant hair cell loss in this region (Fig. 3), therefore we analyzed subcellular structures in P18 mice prior to the loss of hair cells. We quantified the number of stereocilia bundles with typical “U” or “W” bundle shapes (Fig. 4a,b). The mutant mice treated with ASO–29 had significantly fewer atypical bundles in the regions that detect 8 and 16 kHz (P ≤ 0.05), but not 32 kHz (Fig. 4c). This pattern of stereocilia rescue is consistent with the ABR results that demonstrate a rescue of hearing in the 8 and 16 kHz range and less robust rescue in the 32 kHz range. Together, our results indicate a change of bundle structure and number of hair cells at the apical–mid regions, providing an anatomical correlate to function.
Our study demonstrates that a human disease–causing mutation modeled in mice can be corrected to treat deafness and vestibular dysfunction and treatment during the critical hair cell developmental period is likely essential and perhaps sufficient for long-term hearing rescue. Notably, a modest correction of Ush1c splicing is sufficient to rescue hearing in mice for 6 months. This enduring effect of the ASOs is consistent with the duration of action of ASOs observed in the treatment of other mouse models of disease such as spinal muscular atrophy23 and myotonic dystrophy24.
The rescue of hearing in mice using ASO–29 demonstrates that deafness can be treated if intervention occurs early in development. Treatment at P10 corrects vestibular function and partial hearing, whereas treatment at P3–P5 rescues vestibular function and hearing with ABRs comparable to wild–type mice (Supplementary Fig. 1). Although Ush1c is expressed as early as E15 in mice25,26, peak expression in hair cells occurs after P4 and before P16 (https://shield.hms.harvard.edu). Our results are consistent with this expression pattern, suggesting that high expression before P5 is not required for the development of low and mid–frequency hearing, but expression between P5 and P10 may be critical. Hearing at high frequencies (32 kHz) is not rescued to the same level as the lower frequencies, and the rescue is more transient (Fig. 2). Because detection of high frequency sound occurs at the base of the cochlea, this result may suggest that Ush1c is expressed tonotopically during development, and when treated at P5, splicing correction only benefits the mid–apical regions of the cochlea. The development of the ear and hearing in humans occurs in utero27. Thus, treatment in humans would likely require delivery to the fetus via approaches such as intrauterine transfusion28.
Individuals with Usher syndrome suffer a tremendous burden from the dual sensory loss of hearing and vision, and the correction of one of these sensory deficits will have a significant positive impact. Although the RP associated with Usher syndrome is recapitulated in the_Ush1c.216AA_ mice, retinal cell loss occurs later, at approximately one year of life in these mice14. Thus, our analysis of these animals’ vision will require further investigation at later time points. The rescue of hearing in this study offers an important model for studying the development of hearing and vestibular function and for developing approaches to correct these processes when they are impaired.
ONLINE METHODS
Oligonucleotide Synthesis
The synthesis and purification of all 2’O–methoxyethyl-modified oligonucleotides with phosphorothioate backbone and all 5–methyl cytosines, was performed as described29. The oligonucleotides were dissolved in 0.9% saline and stored at –20°C. Sequences are shown in Supplementary Table 1.
Plasmids
The minigene expression plasmid, pCI–Ush1C_216G and 216A were constructed by amplifying genomic DNA from lymphoblast cell lines derived from an _USH1C.216AA_Usher syndrome patient (GM09458, Coriell Institute) or a healthy individual (GM09456, Coriell Institute). PCR primers specific for the 5′ end of exon 2 with restriction sites for XhoI and for the 3′ end of exon 4 with a restriction site for NotI at the 3′ end were used to amplify by PCR the USH1C216A minigene fragment. The PCR product was purified and digested with XhoI and NotI and ligated into the expression plasmid pCI expression vector (Promega) digested with the same restriction enzymes.
Cell culture
Plasmids (1 μg) expressing a minigene of human_USH1C_.216A exons 2–4 and ASOs (50 nM final concentration) were transfected into HeLa cells using Lipofectamine 2000 (Life Technologies). Forty-eight hours after transfection, RNA was isolated using Trizol reagent (Life Technologies) and analyzed by radioactive RT–PCR with primers, pCI FwdB and pCI Rev, to plasmid sequences flanking exon 2 and exon 4.
Mice
Ush1c.216A knock–in mice were obtained from Louisiana State University Health Science Center (LSUHSC)13 and bred and treated at Rosalind Franklin University of Medicine and Science (RFUMS). All procedures met the NIH guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committees at RFUMS and LSUHSC. Mice were genotyped using ear punch tissue and PCR as described previously14. For all studies, both male and female mice were used in approximately equal proportions. For studies in adult mice, homozygous Ush1c.216AA mice (2–4 months of age) were injected intraperitoneally twice a week for two weeks. RNA was isolated from different tissues using Trizol reagent (Life Technologies) and analyzed by radioactive RT–PCR using primers musUSH1Cex2F and musUSH1Cex5F of the Ush1c.216A transgene. Products were separated on a 6% non-denaturing polyacrylamide gel and quantitated using a Typhoon 9400 phosphorimager (GE Healthcare). For studies in neonate mice, pups were injected with 300 mg kg−1 of 2′MOE ASOs at different ages, post-natal day 3–16 (P3–16), as indicated, by intraperitoneal injection. After ABR analysis, animals were euthanized and tissues were collected. For ABR analysis, mice were shipped 2–3 weeks post-treatment to LSUHSC.
Splicing and protein analysis
Retina and inner ears were isolated, cochleae and vestibules separated and immediately frozen in liquid nitrogen or stored in Trizol reagent. For western blot analysis, proteins were obtained from homogenization in a modified RIPA buffer30 or isolated from Trizol reagent (Life Technologies) according to manufacturer’s instructions. Proteins were separated on 4–15% Tris-glycine gradient gels, transferred to membrane and probed with USH1C (#20900002, Novus Biologicals) or β-actin (Sigma Aldrich) specific antibodies. RNA was isolated from different tissues using Trizol reagent (Life Technologies) and analyzed by radioactive RT–PCR using primers musUSH1Cex2F and musUSH1Cex5F of the _Ush1c._216A transgene. Briefly, 0.25–1μg of RNA was reverse transcribed using GoScript Reverse Transcriptase (Promega, Fitchburg, WI) and 1 μl of cDNA was used in PCR reactions with GoTaq Green (Promega) supplemented with primers and 0.1–.25 μl of α–32P–dCTP. Products were separated on a 6% non-denaturing polyacrylamide gel and quantitated using a Typhoon 9400 phosphorimager (GE Healthcare).
Behavioral analysis
Behavioral tests were performed according to previously established protocols15. To quantitate circling behavior, mice were placed in an open-field chamber and behavior was analyzed using ANY-maze behavioral tracking software (Stoelting Co). Ear-twitch, startle and freezing behavior in response to a high amplitude sound was measured by observing mouse activity following a short whistle (Supplementary Fig. 3). Swim tests were performed by placing mice in a tub of room temperature water and observing their swimming behavior for ten seconds. Contact– righting reflex testing was performed by placing the mouse into a closed clear tube or box and measuring the time it took to right when turned upside down. The trunk–curl test was performed by holding the tail and observing whether the mouse reached for a nearby surface or curled toward the base of their tail.
Auditory–evoked brain stem response
Hearing thresholds of treated and untreated Ush1c wt, het and 216AA mutant mice were measured by auditory–evoked brain stem response (ABR). Mice were anesthetized (I.P. ketamine, 100 mg kg−1; xylacine, 6 mg kg−1) and body temperature was maintained near 38°C with a heat pad. All recordings were conducted in a sound proof room. Stimuli consisted of 5 ms pulses of broad–band noise, 8–, 16– and 32 kHz, with 0.5 ms linear ramps. Although these tone stimuli encompass low, medium and high regions of mouse spectral sensitivity, BBN was included to confirm that responses are representative of the whole cochlear response. The stimuli were broadcast through a Motorola piezoelectric speaker (Model No. 15D87141E02) fitted with a plastic funnel and 2 mm diameter tubing over the speaker front, producing an acoustic wave guide which was positioned in the external meatus approximately 0.5 cm from the tympanum. Using continuous tones, stimulus amplitude was calibrated at the end of the tubing with a Bruel and Kjaer 2610 measuring amplifier (fast, linear weighting), 4135 microphone (grid on) and 4230 pistonphone calibrator. All stimulus amplitudes were dB SPL (re. 20 μPa). Total harmonic distortion was –40 dB (Hewlet Packard 3562A Signal Analyzer). Stimuli were generated (195 kHz srate) and responses digitized (10 kHz srate) using TDT System III (TDT) hardware and software (BioSig). ABRs were recorded with a silver wire (0.03 o.d.) placed subcutaneously behind the left ear, with indifferent and ground electrodes (steel wire) placed subcutaneously at the vertex and hind–limbs, respectively. Responses to 5 msec broad-band noise, 8–, 16–, and 32–kHz tone bursts were recorded. After amplification (60 dB, Grass P5 AC), filtering (0.3 Hz–1 kHz; TDT PF1), and averaging (n = 600–1024), thresholds (+/− 6 dB) were determined by eye as the minimum stimulus amplitude which produced an ABR wave pattern similar to that produced for the highest intensity stimulus (90 dB).
Scanning Electron Microscopy (SEM)
The SEM analysis was performed as has been previously described14. Specifically, intra-labyrinthine perfusion with 2.5% glutaraldehyde/1% paraformaldehyde/1.5% sucrose in 0.12 M phosphate buffer (pH 7.4) was performed on whole cochleae dissected from mice at P18. Cochleae were post-fixed by immersion in for 1 day in the same fixative at 4°C with gentle rotation followed by three washes in 0.12 M phosphate buffered saline (PBS) and stored for one week at 4°C. Cochleae were next fixed in 1% OsO4 in PBS for 40 min and washed in PBS. Specimens were then serially dehydrated in ethanol, dried in a critical point drier (Autosamdri-814, Tousinis Research Corporation), and mounted on aluminum stubs. The bony capsule of the cochlea, spiral ligament, stria vascularis, and Reissner’s membrane were removed and the whole organ of Corti was exposed with fine dissecting instruments. Specimens were coated in gold/palladium with a Hummer VIA sputter coater (Anatech) and viewed on a JEOL JSM 6300 F scanning electron microscope. At least three individual animals representative of each experimental paradigm were analyzed.
The cochlear place–frequency map relating distance from cochlear apex and frequency is based on a tonotopic map of mice with an average basilar membrane distance from apex to base of 5.13 mm21. Distances were calculated using the equation: d = 156.5 - 82.5 × log(f); d is the normalized distance from the base (%) and f, frequency in kHz21.
Immunofluorescence
Fluorescent labeling of microdissected preparations of the organ of Corti were used to study the hair cells of one month old treated and untreated mutant and control mice as described previously14,31. Briefly, cochleae were isolated from the auditory bulla and a small opening was created in the apex. The stapes was removed from the oval window and the cochleae were gently perfused with 2% paraformaldehyde in 0.1M phosphate buffer, pH 7.4 and post-fixed by immersion for 2 hours at 4°C with gentle rocking. Tissues were washed twice with PBS following fixation and processed for immunohistochemistry. For harmonin analysis, the tectoral membrane was removed with a fine forceps and the stria vascularis was trimmed. Tissues were blocked for 1 hour at room temperature or overnight at 4°C (harmonin analysis) in a blocking solution consisting of 10% normal donkey serum, 0.5% bovine serum albumin, 0.1% Triton X– 100, and 0.03% saponin in PBS in order to reduce non–specific binding of primary and secondary antibodies. Primary antibody incubations were then performed at 4°C in PBS containing 5% normal donkey serum, 0.05 % bovine serum albumin, 0.1% Triton X-100, and 0.03% saponin in PBS. For counting cells, a mouse monoclonal anti-parvalbumin antibody (parv19, Cat. No. P3088, Sigma, 1:250) was used to label cochlear hair cells32. To analyze harmonin b expression, polyclonal rabbit anti-harmonin antibodies specific to isoform b (gift from Uwe Wolfrum, 1:100) were used. For mouse antibodies against parvalbumin, the M.O.M. kit was used as specified by the manufacturer (Vector Labs). Tissues were washed (3 times for 10-15 min. each) after primary and secondary antibody (Donkey anti-mouse Alexa555 and Donkey anti-rabbit Alexa488, 1:400, Invitrogen) incubations in 0.1% Tween-20 in PBS and nuclei were counterstained with DAPI (1 μg/ml; Cat. No. D9542, Sigma-Aldrich). F-actin was labeled with rhodamine phalloidin (Life Technologies) according to the manufacturer’s instructions. For counting hair cells, specimens were dehydrated through an ethanol series, cleared with methyl salicylate:benzl benzoate (5:3) and examined by confocal fluorescence microscopy. For harmonin b analysis, labeled specimens were mounted and stored in Prolong Gold (Invitrogen). All samples were imaged with a Zeiss motorized system operated with LSM software (Zeiss) and equipped with 405, 543, and 633nm diodes along with a multi-line argon laser (457nm, 488nm, 515nm); an XYZ stage; and several objectives that include the Plan-NEOFLUAR 10x (NA=0.3), Plan-NEOFLUAR 40x (AN=1.3 oil) and Plan-APOACHROMAT100x (NA=1.4 oil) used. Scans were performed through a sequential (line) mode and PMT voltages dynamically regulated to compensate for signal loss due to scatter and depth limitations. Planes were captured at a resolution of 2048x2048 and speeds of 20-200 us/pixel. Optical volumes were deconvolved with a constrained maximum likelihood estimation algorithm and a calculated point spread function using Huygens Professional 4.1 (Scientific Volume Imaging) running on a Mac Pro computer (Apple). Z stack images were reconstructed and analyzed using ImageJ, Fuji and Photoshop softwares.
Statistics
Data were analyzed by ANOVA with post hoc tests and Student’s t-test (SAS Institute Inc, NC or Prism 5 Graphpad Software) as noted in the Figure legends. Hair cell counts were analyzed as the dependent variable separately for both inner and outer hair cells in a nested analysis of variance with a two level factorial arrangement of treatments33. The nested effect was the mice within each genotype treatment combination, the two main effect factors were cell location in the cochlea and genotype/treatment combination (combined into one variable with three levels, see Fig. 4). Adjustment for multiple comparisons conducted to separate interaction means was by a simulation method34. All data management and analysis was performed using programs and procedures in the Statistical Analysis System (SAS Institute).
Supplementary Material
1
2
3
Acknowledgements
We gratefully acknowledge support from the Hearing Health Foundation, Midwest Eye–Banks, Capita Foundation, and the National Institutes of Health. We thank D. Cunningham and E. Rubel for assistance with SEM analysis; A. Rosenkranz, R. Marr, and M. Oblinger for use of equipment; J. Huang for assistance with open–field analysis, L. Ochoa for assistance with ABR analysis computer support, G. MacDonald with confocal imaging and deconvolution analysis, U. Wolfrum (Johannes Gutenberg University of Mainz, Germany) for harmonin b antibody, H. Thompson for statistical analysis, and A. Case, B. Keats and M. Havens for discussions and comments on the manuscript.
Footnotes
Author Contributions The project was conceived by M.L.H. Experiments were designed and performed by A.J.H, F.M.J, J.J.L., K.E.M., M.L.H., and D.M.D. and were analyzed by M.L.H., J.J.L., F.R., F.M.J, A.J.H., and K.E.M. Animal work was carried out by M.L.H., D.M.D., K.E.M. A.J.H., F.M.J., J.J.L., M.J.S. Molecular experiments were performed by A.J.H., F.M.J, K.E.M., and M.L.H. J.J.L. carried out the immunofluorescence analysis. J.J.L, M.J.S. and H.E.F. performed auditory brainstem response experiments and J.J.L. and N.G.B. interpreted the results. M.L.H. and J.J.L wrote the paper.
Competing financial interests F.R. may materially benefit either directly or indirectly through stock options. M.L.H. could materially benefit if a therapeutic for Usher results from this work.
Treatment of deafness using antisense oligonucleotides
REFERENCES
- 1.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Dror AA, Avraham KB. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009;43:411–437. doi: 10.1146/annurev-genet-102108-134135. [DOI] [PubMed] [Google Scholar]
- 3.de Felipe MM, Feijoo Redondo AF, Garcia-Sancho J, Schimmang T, Alonso MB. Cell- and gene-therapy approaches to inner ear repair. Histol Histopathol. 2011;26:923–940. doi: 10.14670/HH-26.923. [DOI] [PubMed] [Google Scholar]
- 4.Di Domenico M, et al. Towards gene therapy for deafness. J Cell Physiol. 2011;226:2494–2499. doi: 10.1002/jcp.22617. [DOI] [PubMed] [Google Scholar]
- 5.Bermingham-McDonogh O, Reh TA. Regulated reprogramming in the regeneration of sensory receptor cells. Neuron. 2011;71:389–405. doi: 10.1016/j.neuron.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitner-Glindzicz M, et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet. 2000;26:56–60. doi: 10.1038/79178. [DOI] [PubMed] [Google Scholar]
- 7.Verpy E, et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 8.Kimberling WJ, et al. Frequency of Usher syndrome in two pediatric populations: Implications for genetic screening of deaf and hard of hearing children. Genet Med. 2010;12:512–516. doi: 10.1097/GIM.0b013e3181e5afb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang XM, et al. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum Genet. 2005;116:292–299. doi: 10.1007/s00439-004-1227-2. [DOI] [PubMed] [Google Scholar]
- 10.Ebermann I, et al. Deafblindness in French Canadians from Quebec: a predominant founder mutation in the USH1C gene provides the first genetic link with the Acadian population. Genome Biol. 2007;8:R47. doi: 10.1186/gb-2007-8-4-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang XM, et al. USH1C: a rare cause of USH1 in a non-Acadian population and a founder effect of the Acadian allele. Clin Genet. 2003;63:150–153. doi: 10.1046/j.0009-9163.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 12.Lentz J, et al. The USH1C 216G-->A splice-site mutation results in a 35-base-pair deletion. Hum Genet. 2005;116:225–227. doi: 10.1007/s00439-004-1217-4. [DOI] [PubMed] [Google Scholar]
- 13.Lentz J, Pan F, Ng SS, Deininger P, Keats B. Ush1c216A knock-in mouse survives Katrina. Mutat Res. 2007;616:139–144. doi: 10.1016/j.mrfmmm.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Lentz JJ, et al. Deafness and retinal degeneration in a novel USH1C knock-in mouse model. Dev Neurobiol. 2010;70:253–267. doi: 10.1002/dneu.20771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardisty-Hughes RE, Parker A, Brown SD. A hearing and vestibular phenotyping pipeline to identify mouse mutants with hearing impairment. Nat Protoc. 2010;5:177–190. doi: 10.1038/nprot.2009.204. [DOI] [PubMed] [Google Scholar]
- 16.Michalski N, et al. Harmonin-b, an actin-binding scaffold protein, is involved in the adaptation of mechanoelectrical transduction by sensory hair cells. Pflugers Arch. 2009;459:115–130. doi: 10.1007/s00424-009-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grillet N, et al. Harmonin mutations cause mechanotransduction defects in cochlear hair cells. Neuron. 2009;62:375–387. doi: 10.1016/j.neuron.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefevre G, et al. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 2008;135:1427–1437. doi: 10.1242/dev.012922. [DOI] [PubMed] [Google Scholar]
- 19.Boeda B, et al. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng AW, Salles FT, Pan B, Ricci AJ. Integrating the biophysical and molecular mechanisms of auditory hair cell mechanotransduction. Nat Commun. 2011;2:523. doi: 10.1038/ncomms1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller M, von Hunerbein K, Hoidis S, Smolders JW. A physiological place-frequency map of the cochlea in the CBA/J mouse. Hear Res. 2005;202:63–73. doi: 10.1016/j.heares.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Greenwood DD. A cochlear frequency-position function for several species--29 years later. J Acoust Soc Am. 1990;87:2592–2605. doi: 10.1121/1.399052. [DOI] [PubMed] [Google Scholar]
- 23.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Amraoui A, Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- 26.Petit C, Richardson GP. Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci. 2009;12:703–710. doi: 10.1038/nn.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall JW., 3rd Development of the ear and hearing. J Perinatol. 2000;20:S12–20. doi: 10.1038/sj.jp.7200439. [DOI] [PubMed] [Google Scholar]
- 28.Uhlmann RA, Taylor M, Meyer NL, Mari G. Fetal transfusion: the spectrum of clinical research in the past year. Curr Opin Obstet Gynecol. 2010;22:155–158. doi: 10.1097/GCO.0b013e32833723b5. [DOI] [PubMed] [Google Scholar]
- 29.Baker BF, et al. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhe ion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J Biol Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 30.Hastings ML, et al. Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci Transl Med. 2009;1:5ra12. doi: 10.1126/scitranslmed.3000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardie NA, MacDonald G, Rubel EW. A new method for imaging and 3D reconstruction of mammalian cochlea by fluorescent confocal microscopy. Brain Res. 2004;1000:200–210. doi: 10.1016/j.brainres.2003.10.071. [DOI] [PubMed] [Google Scholar]
- 32.Sage C, Venteo S, Jeromin A, Roder J, Dechesne CJ. Distribution of frequenin in the mouse inner ear during development, comparison with other calcium-binding proteins and synaptophysin. Hear Res. 2000;150:70–82. doi: 10.1016/s0378-5955(00)00183-0. [DOI] [PubMed] [Google Scholar]
- 33.Milliken GA, Johnson DE. Analysis of Messy Data Volume I: Designed Experiments. Lifetime Learning Publications; Belmont, CA: 1984. [Google Scholar]
- 34.Edwards D, Berry JJ. The efficiency of simulation-based multiple comparisons. Biometrics. 1987;43:913–928. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3