Ultraviolet Radiation Damages Self Noncoding RNA And Is Detected By TLR3 (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 30.
Published in final edited form as: Nat Med. 2012 Jul 8;18(8):10.1038/nm.2861. doi: 10.1038/nm.2861
Abstract
Exposure to ultraviolet B (UVB) radiation from the sun can result in sunburn, premature aging and carcinogenesis, but the mechanism responsible for acute inflammation of the skin is not well understood. Here we show that RNA is released from keratinocytes after UVB exposure and that this stimulates production of the inflammatory cytokines tumor necrosis factor α (TNF-α) and interleukin-6 (IL-6) from nonirradiated keratinocytes and peripheral blood mononuclear cells (PBMCs). Whole-transcriptome sequencing revealed that UVB irradiation of keratinocytes induced alterations in the double-stranded domains of some noncoding RNAs. We found that this UVB-damaged RNA was sufficient to induce cytokine production from nonirradiated cells, as UVB irradiation of a purified noncoding RNA (U1 RNA) reproduced the same response as the one we observed to UVB-damaged keratinocytes. The responses to both UVB-damaged self-RNAs and UVB-damaged keratinocytes were dependent on Toll-like receptor 3 (TLR3) and Toll-like receptor adaptor molecule 1 (TRIF). In response to UVB exposure, _Tlr3_−/− mice did not upregulate TNF-α in the skin. Moreover, TLR3 was also necessary for UVB-radiation–induced immune suppression. These findings establish that UVB damage is detected by TLR3 and that self-RNA is a damage-associated molecular pattern that serves as an endogenous signal of solar injury.
Excess exposure to solar radiation in the UVB wavelength range (280–320 nm) is a key risk factor for skin cancer1,2 and results in an inflammatory reaction of the skin commonly known as sunburn. However, despite the morbidity resulting from inflammation caused by UVB radiation, the molecular mechanisms responsible for detecting UVB injury are not completely understood. Once UVB damage occurs, several downstream responses have been described, including changes in _cis_-urocanic acid (_cis_-UCA), DNA and lipids3–5. UVB exposure also triggers the activation of nuclear factor-κ B (NF-κB)6,7 and the induction of cytokines, including IL-6 and TNF-α8. TNF-α is thought to have a role in the response to UVB damage, as it induces apoptosis9, is proinflammatory10,11 and may mediate UVB immunosuppression12,13, and TNF-α–specific antibodies reduce the number of sunburn cells after UVB exposure14. Because of the major pleiotropic, downstream effects of TNF-α after solar injury, we sought here to determine what elements induce the expression of this cytokine after exposure to UVB radiation.
We first determined if exposing cells to UVB radiation produced a molecule(s) capable of inducing the release of TNF-α from non-irradiated cells. We exposed normal human epidermal keratinocytes (NHEKs) to UVB radiation (15 mJ cm−2) and added the lysates from the UVB-irradiated keratinocytes or equal numbers of lysed non-irradiated cells to nonirradiated NHEKs and PBMCs. Lysates from UVB-irradiated NHEKs stimulated the production of TNF-α and IL-6 in nonirradiated, responding NHEKs and PBMCs, but lysates from nonirradiated NHEKs did not (Fig. 1a–c). The concentrations of TNF-α protein remained elevated at up to 48 h after exposure to UVB-irradiated keratinocyte lysates (Supplementary Fig. 1). RNase inhibited the capacity of the lysates from UVB-irradiated NHEKs to induce the production of TNF-α and IL-6 (Fig. 1a–c). We found that the lysates from UVB-irradiated NHEKs also induced an increase in the expression of TLR3 in responding nonirradiated NHEKs (Fig. 1d), a pattern recognition molecule that is best known for its capacity to detect dsRNA15. Targeted knockdown of TLR3 in nonirradiated responding NHEKs significantly (P < 0.001) reduced the capacity of lysates from UVB-irradiated NHEKs to induce the production of TNF-α and IL-6 (Fig. 1e,f). We were also able to reproduce these events in mice, as intradermal injection of lysates from UVB-irradiated NHEKs, but not of nonirradiated NHEKs, induced redness, swelling and TNF-α and IL-6 cytokine production in the skin of wild-type mice but not the skin of _Tlr3_−/− mice (Fig. 1g–i and Supplementary Fig. 2). Thus, these observations suggest that recognition of UVB-induced damage may be mediated by RNA and detected by TLR3.
Figure 1.
RNA from UVB-irradiated keratinocytes induces the production of inflammatory cytokines. (a) Quantitative PCR (qPCR) measurements of TNF-α and IL-6 mRNA (24 h) in human keratinocytes cultured for 24 h with control NHEK lysates (nonirradiated), lysates from UVB-irradiated (UVR) NHEKs or lysates from UVR NHEKs first treated with RNase. (b) The concentrations of TNF-α and IL-6 protein measured by ELISA from culture supernatants of NHEKs after the additions described in a. (c) The concentrations of TNF-α and IL-6 protein from PBMCs treated with lysates from UVR NHEKs as described in a. (d,e) qPCR measurements of TLR3 mRNA (24 h) (d) and of TNF-α and IL-6 mRNA (e) from NHEKs treated with TLR3 siRNA or control NHEKs treated with vehicle, transfection reagent (DF) or control oligonucleotides (ctrl siRNA) and then stimulated with lysates from UVR NHEKs. (f) Concentrations of TNF-α and IL-6 protein from culture supernatant of NHEKs measured by ELISA after the experiments described in e. (g) The ears of a wild-type C57BL/6 mouse and a _Tlr3_-/- mouse 24 h after intradermal ear injection of lysates from UVR NHEKs or equal amounts of lysates from nonirradiated NHEKs. (h) Thickness of the ear skin in mice treated as described in g. (i) qPCR measurements of TNF-α and IL-6 mRNA in tissue extracts of ear skin treated as described in g. To determine statistical significance between groups, comparisons were made using two-tailed t tests *P < 0.05, ***P < 0.001. Data are means ± s.e.m. and are representative of at least three independent experiments. n = 4–6 mice per group. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
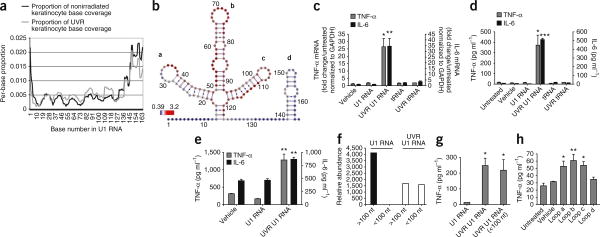
To identify specific alterations in RNA induced by UVB exposure that may be responsible for triggering the inflammatory response, we applied an unbiased approach using an adaptation of next-generation whole-transcriptome shotgun sequencing (RNA-Seq). This method of RNA sequencing can be dependent on the accessibility of the RNA secondary structure, and a change in RNA structure induced by UVB radiation can therefore be detected by a change in the frequency of sequence reads. A comparison of the base-reading frequency obtained using RNA from lysates from UVB-irradiated NHEKs and that obtained using RNA from identical unexposed cells revealed changes in the specific domains of several small nuclear RNAs (snRNAs) (Supplementary Table 1). These snRNAs were of particular interest because they contain stem-loop structures that could form dsRNAs and thereby activate TLR3. We took particular note of snU1 RNA, as it was among the most abundant noncoding RNAs present in the irradiated cells (falling into the ninety-fourth percentile), and its sequence frequency increased by 19.5% after UVB exposure (Supplementary Table 1). An RNA-Seq analysis of the U1 RNA showed that UVB exposure increased the base-read frequency in the U1 loop domains (Fig. 2a,b). These loops generated were potential ligands for TLR3.
Figure 2.
UVB damage to U1 RNA generates products that induce the production of TNF-α and IL-6. (a) Base-read frequency of specific domains of U1 RNA from keratinocytes after exposure to 15 mJ cm−2 UVB, as determined by whole-transcriptome RNA sequencing (RNA-Seq). Data are shown with the per-base coverage as a proportion of the total sequencing coverage. (b) Representation of a sequencing analysis by base coverage showing that the loop domains a, b and c in U1 RNA increase in frequency after UVB exposure (red), whereas loop d and the liner domains decrease in frequency (blue). Numbers around the diagram indicate the base number in U1 RNA. (c) qPCR measurements of TNF- α and IL-6 mRNA in NHEKs 24 h after the addition of 100 ng of UVB irradiation (UVR) (15 mJ cm−2) U1 RNA. The addition of 100 ng of tRNA did not stimulate the NHEKs. (d) The amount of TNF-α and IL-6 protein released by NHEKs into the media 24 h after the addition of 100 ng of UVR (15 mJ cm−1) U1 RNA. (e) The amount of TNF-α and IL-6 protein released by PBMCs into the media 24 h after the addition of 100 ng of UVR (15 mJ cm−1) U1 RNA. (f) The relative abundance of U1 RNA detected at sizes greater than and less than 100 nt as determined by size exclusion gel purification before and after exposure to 15 mJ cm−2 UVB radiation. (g) The concentration of TNF-α protein in the NHEK media 24 h after the addition of UVB-generated fragments of U1 RNA less than 100 nt in length. (h) TNF-α mRNA (24 h) in NHEKs after treatment with synthetic oligonucleotides based on loops a–d of U1 RNA. To determine statistical significance between groups, comparisons were made using two-tailed t tests *P < 0.05, **P < 0.01, ***P < 0.001. Data are means ± s.e.m. and are representative of at least three independent experiments.
As U1 RNA had been suggested to modulate autoimmune responses and activate innate immune signaling16,17, we chose to test whether direct UVB damage to this type of endogenous noncoding RNA could trigger an inflammatory response. We exposed pure, synthetic U1 RNA to UVB radiation and then added it to NHEKs or PBMCs. Nonirradiated U1 RNA (3,000 ng ml−1) had no detectable effect on inducing the production of TNF-α in either cell type (Fig. 2c–e). In contrast, low concentrations (100 ng ml−1) of U1 RNA that was exposed to UVB irradiation increased the production of TNF-α and IL-6 in both NHEKs and PBMCs at 24 h after exposure (Fig. 2c–e). The concentrations of TNF-α remained elevated up to 72 h after exposure in NHEKs (Supplementary Fig. 1).
To confirm that UVB irradiation altered the U1 RNA, we performed a direct analysis of synthetic U1 RNA before and after exposure to UVB radiation. Gel purification of the U1 RNA showed that UVB irradiation resulted in the production of small RNA fragments less than 100 bp in length (Fig. 2f). We obtained a similar result after an analysis of UVB-irradiated U1 RNA with capillary electrophoresis (data not shown). These purified fragments of U1 RNA generated by UVB irradiation potently stimulate TNF-α production when added to nonirradiated NHEKs (Fig. 2g). Furthermore, small synthetic oligonucleotides representing the U1 RNA loops a, b and c (Fig. 2b) induced TNF-α production when added to nonirradiated NHEKs, but oligonucleotides representing loop d did not show activity in this context (Fig. 2h). Thus, activity of the oligonucleotides correlated with the over-representation of loops a–c and the under-representation of loop d found by RNA-Seq analysis.
We next examined whether U1 RNA could activate TLR3 signaling. As observed with total RNA from UVB-damaged cells, UVB-irradiated U1 RNA significantly (P < 0.05) induced the expression of TLR3 mRNA in cultured nonirradiated NHEKs (Fig. 3a). Treatment of NHEKs with TLR3 siRNA attenuated the response to UVB-irradiated U1 RNA (Fig. 3b,c). Confocal microscopy and intracellular staining for TLR3 confirmed that UVB-irradiated U1 RNA interacts with endosomal TLR3 in NHEKs (Fig. 3d). UVB-irradiated U1 RNA also induced the translocation of the transcription factor RelA (also known as p65) to the nucleus, as assayed by western blot (Fig. 3e), and this was dependent on TLR3, as assayed by direct immunofluorescence (Fig. 3f). Furthermore, pretreatment of NHEKs for 2 h with either dextran sulfate or fucoidan, two inhibitors of scavenger receptor function and dsRNA uptake, inhibited the TNF-α–inducing activity of the UVB-irradiated U1 RNA, suggesting uptake and endosomal delivery of U1 RNA is required to activate TLR3 (Supplementary Fig. 3).
Figure 3.
UVB damage to U1 RNA induces inflammatory cytokine release by activating TLR3. (a) TLR3 mRNA expression in NHEKs measured by qPCR after 24 h of culture with UVB-irradiated (UVR) U1 RNA. (b) TNF-α and IL-6 mRNA expression from NHEKs treated with siRNA to TLR3 and then stimulated with UVR U1 RNA for 24 h. (c) The concentrations of TNF-α and IL-6 in supernatants from NHEKs treated with siRNA to TLR3. (d) Intracellular colocalization of TLR3 (red fluorescence) and UVR U1 RNA labeled with Alexa Fluor 488 (green fluorescence). The blue staining is DAPI. Scale bars, 20× magnification, 50 μm; 100× magnification, 20 μm. (e) RelA and lamin B1 (loading control) detected by western blot of nuclear lysates of NHEKs treated with UVR U1 RNA for 1, 2 or 4 h. (f) Results from NHEKs cultured to 70% confluency on chamber slides, treated with siRNA to TLR3 and exposed to UVR U1 RNA for 4 h. Cells were fixed and stained with an antibody to RelA (red). Scale bar, 50 μm. To determine statistical significance between groups, comparisons were made using two-tailed t tests *P < 0.05, **P < 0.01, ***P < 0.001. Data are means ± s.e.m. and are representative of at least three independent experiments.
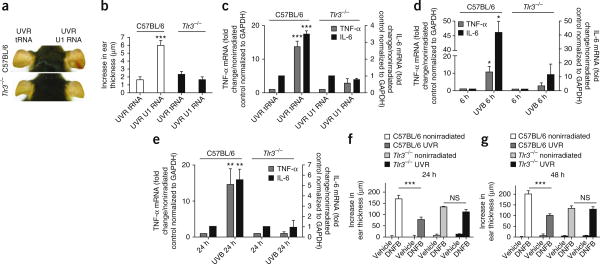
Next, we assessed the effects of U1 RNA in vivo. UVB-irradiated U1 RNA induced redness and increased ear thickness in mouse ears, whereas transfer RNA (tRNA) exposed to UVB did not (Fig. 4a,b). UVB-irradiated U1 RNA also increased the expression of TNF-α and IL-6 mRNA in the skin of wild-type control but not _Tlr3_−/− mice (Fig. 4a–c). Furthermore, the inflammatory response to UVB-exposed U1 RNA was blunted in mice deficient in TRIF, an essential downstream element in TLR3 signaling (Supplementary Fig. 4).
Figure 4.
Recognition of UVB-irradiated RNA by TLR3 is necessary for the inflammatory response to UVB damage. (a) The ears of a wild-type C57BL/6 mouse and a _Tlr3_-/- mouse 24 h after intradermal ear injection of UVB-irradiated (UVR) U1 RNA or tRNA. (b) Thickness of the ear skin treated as described in a. (c) qPCR measurements of TNF-α and IL-6 mRNA in tissue extracts of ear skin treated as described in a.(d,e) TNF-α and IL-6 mRNA expression in the skin of wild-type C57BL/6 and _Tlr3_-/- mice 6 h (d) and 24 h (e) after exposure to 5 kJ m−2 UVB. (f,g) Ear swelling 24 h (f) and 48 h (g) after DNFB challenge (Online Methods). To determine statistical significance between groups, comparisons were made using two-tailed t tests, NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001. Data are means ± s.e.m. and are representative of at least three independent experiments. n = 4–6 mice per group.
To assess the role of TLR3 in the response to UVB radiation, we directly irradiated mice and measured both the inflammatory response and UVB-radiation–induced immunosuppression. Six hours and 24 h after UVB exposure, upregulation of TNF-α or IL-6 mRNA in the skin was reduced in _Tlr3_−/− mice as compared to wild-type littermates (Fig. 4d,e). In addition, UVB exposure did not induce the production of inflammatory cytokines in _Trif_−/− mice (Supplementary Fig. 4). _Tlr3_−/− mice also showed less skin redness compared to wild-type controls after UVB radiation exposure (Supplementary Fig. 5). Although IL-1 stimulates the production of TNF-α and has a key role in inflammation after UVB exposure18,19, TNF-α was not induced in the skin of IL-1 receptor knockout mice (_Il1r_−/−) after UVB exposure (Supplementary Fig. 6). TLR3 was also necessary for UVB-radiation– induced immunosuppression after UVB exposure. Twenty-four hours after sensitization and challenge with dinitrofluorobenzene (DNFB), nonirradiated wild-type mice developed an allergic contact hyper-sensitivity (CHS) reaction (Fig. 4f,g). Exposure to UVB radiation 72 h before sensitization to DNFB suppressed the CHS response in wild-type but not in _Tlr3_−/− mice (Fig. 4f,g and Online Methods), suggesting that TLR3 is crucial for the ability of UVB radiation to suppress the CHS response.
The recognition of RNA involves a coordinated response between TLRs and cytoplasmic RNA helicases. In our studies, however, there was little indication of any involvement of other RNA sensors. The induction of TNF-α and IL-6 were not altered after UVB exposure of mice deficient in mitochondrial antiviral signaling protein (MAVS), the adaptor for intracellular RNA helicases such as melanoma differentiation-associated gene 5 (MDA5) and retinoic acid inducible gene-1 (RIG-1) (Supplementary Fig. 7). Furthermore, the use of a pharmacologic inhibitor of this pathway did not block the induction of cytokines by lysates from UVB-irradiated NHEKs (Supplementary Fig. 7).
Based on these observations, we propose that noncoding RNAs damaged by UVB radiation are partially responsible for UVB-induced inflammation and act as previously unknown damage-associated molecular patterns. In support of our hypothesis are prior observations that UVB radiation can induce RNA-RNA crosslinking, pyrimidine dimer formation and the oxidation of guanine to 8-oxo-7,8-dihydroguanosine (8-oxoG)20. Other molecules may also activate TLR3 in the whole-cell system, but we here show specifically that a specific form of RNA altered by UVB irradiation can activate TLR3, as isolated tRNA and nonirradiated U1 RNA were inactive in this context. In addition, other products generated after UV exposure, such as _cis_-UCA21, telomeric nucleotides22,23, 6-formylindolo(3,2-b) carbazole24 and IL-1 (refs. 25,26), may also contribute to UVB-induced inflammation but probably do not contribute substantially to the immediate production of TNF-α and IL-6, as this response was dependent on TLR3.
These observations provide a new framework to understand the UVB damage response and may influence the understanding of a wide range of photosensitive phenomena. They suggest the potential for RNAs as biomarkers of UVB-induced injury, the use of RNAs as alternatives to phototherapy and the potential for blocking damaged self-RNA recognition to be used as a therapeutic target for photosensitive disorders. The TLR3-mediated cytokine response may have major implications for tumorigenesis because of the many associations of TNF-α with apoptosis, inflammation and immune surveillance. Future work to better define the role of TLR3 in the outcomes occurring after UVB exposure is therefore warranted.
Online Methods
Cell culture
NHEKs were grown in serum-free EpiLife cell culture media (Cascade Biologics) containing 0.06 mM Ca2+ and EpiLife Defined Growth Supplement at 37 °C under standard tissue culture conditions. The cultures were maintained for up to four passages in this media, with the addition of 50 U ml−1 of penicillin and 50 μg ml−1 of streptomycin. Cells were treated at 70–80% confluence. Human PBMCs were prepared by Ficoll density gradient separation.
Mice
Sex-matched C57BL/6 wild-type controls, male and female TLR3-deficient, male and female TRIF-deficient, female MAVS-deficient and female IL-1R–deficient mice on a C57BL/6 background were housed at the University Research Center at the University of California, San Diego (UCSD). All animal experiments were approved by the UCSD Institutional Animal Care and Use Committee. Mice were administered intradermal ear injections of UVB-irradiated keratinocytes (1.2 × 105) or UVB-irradiated U1 RNA (1 μg) (described below). Ear thickness was measured using a micrometer. Six-millimeter punch biopsies were performed after CO2 euthanasia to harvest tissue for histological, mRNA and protein analyses.
Contact Hypersensitivity Experiments
Mice were exposed to UVB (1.44 kJ m−2). Seventy-two hours after the last treatment of UVB, mice were sensitized with 0.5% of DNFB on the irradiated site. Six days after the sensitization, mice were challenged with 0.1% DNFB. Ear thickness was measured at 24 h and 48 h after the challenge.
UVB Exposure
NHEKs were irradiated with UVB at 15 mJ cm−2, as previously described27. Dosimetry was done using a digital ultraviolet radiometer by Solartech Inc. UVB-irradiated cells were used immediately after exposure, and lysates from 600,000 cells were added to 200,000 NHEKs grown to 80% confluence or 200,000 PBMCs in culture dishes. For the mouse injection experiments, 1.2 × 105 cells were used. Sonicated nonirradiated NHEKs treated identically were used as controls. The amounts of TNF-α and IL-6 were measured in the culture media, and mRNA expression was measured in the treated cells 24 h after the addition of lysates from the UVB-irradiated cells. For mouse irradiation, hair was plucked from the back, and 24 h later, the hairless skin was exposed to UVB (5 kJ m−2). The skin was biopsied 6 h and 24 h after irradiation for cytokine measurement.
RNA-Seq
Sequencing libraries were prepared using double-stranded complementary DNA (cDNA) produced using NuGEN RNASeq Ovation kits and 100 ng of total RNA starting material for each sample following the manufacturer's protocols. One-hundred nanograms of double-stranded cDNA was digested with 50 units of S1 nuclease (Promega) for 30 min at 37 °C in 50 mM sodium acetate (pH 4.5), 280 mM NaCl and 4.5 mM ZnSO4. The cDNA library was purified using a DNA Clean&Concentrator-5 Kit (Zymo Research Corp). DNA ends were repaired using 15 units of T4 DNA polymerase (Enzymatics, Beverly, MA), 5 units of Klenow Large Fragment (Enzymatics) and 50 units of T4 polynucleotide kinase (Enzymatics) at 20 °C for 30 min in a mixture of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM ATP, 10 mM dithiothreitol and 400 u\M dNTP. DNA products were purified again using a DNA Clean&Concentrator-5 Kit. Next, DNA ends were A-tailed with 15 units of Klenow (3′→5′ exo-) (Enzymatics) at 37 °C for 30 min in 10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol and 0.2 mM dATP. DNA products were again purified using the DNA Clean&Concentrator-5 Kit. Next, Illumina paired-end adaptor oligonucleotides, at a concentration of 2 u\M, were ligated to the A-tailed cDNA ends with 3,000 units of T4 DNA ligase (Enzymatics) at 20 °C for 15 min in 66 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP and 1 mM polyethyl-eneglycol. DNA products were purified using a DNA Clean&Concentrator-5 Kit. The DNA library products were then separated on a 2% NuSieve GTG agarose gel (Lonza), and products corresponding to a size of approximately 200-300 bases were excised from the gel and isolated using a Zymoclean Gel DNA Recovery Kit (Zymo Research Corp). The excised DNA material was PCR amplified with 1 unit of Phusion Polymerase (Finnzymes) in standard 1× Phusion HF buffer with 0.2 mM dNTPs and 0.6 u\M of the PCR primers PE 1.0 and PE 2.0 (Illumina) for 15 cycles. The amplified DNA products were further purified on 2% NuSieve GTG agarose gel (Lonza), excised and isolated again using a Zymoclean Gel DNA recovery kit. The purified DNA library was quantified using the Qubit quantification platform (Invitrogen) and sized using the 2100 Bioanalyzer (Agilent). DNA products were then denatured in 0.1 N NaOH and diluted to a final concentration of 10 pM before being loaded onto the Illumina paired-end flow cell for massively parallel sequencing by synthesis on the Illumina GAIIx.
Confocal Microscopy
NHEKs were treated with biotinylated UVB-irradiated U1 RNA for 8 h. Cells were fixed and stained using an intracellular TLR staining kit from Imgenex. Cells were incubated with a mouse IgG TLR3-specific Alexa Fluor 647 antibody (1:500 dilution, Imgenex, 40C1285.6) or a mouse IgG isotype control antibody (Imgenex, MOPC-31C) and an Avidin Alexa Fluor 488 conjugate for 1 h. Cells were photographed with a confocal microscope (Zeiss LSM 5 Pascal).
Statistical Analyses
To determine statistical significance between groups, comparisons were made using two-tailed t tests. Analyses of multiple groups were done by one-way or two-way analysis of variance with a Bonferroni post hoc test in GraphPad Prism Version 4. For all statistical tests, P < 0.05 was accepted as statistically significant. All experiments, excluding the RNA-Seq experiments, were repeated at least three times. RNA-Seq was performed twice.
Additional methods
Detailed methodology is described in the Supplementary Methods.
Supplementary Material
supp figures
Acknowledgments
We thank S. Head from The Scripps Research Institute DNA Core Facility for performing RNA-Seq. We thank B. Gilchrest (Boston University, Boston, MA) for telomere oligonucleotides and advice, J. Laskin for advice and helpful discussion and M. Karin (University of California, San Diego, San Diego, CA) for providing _Il1r_−/mice and for helpful discussion. This work was supported by US National Institutes of Health (NIH) grants R01-AR052728, NIH R01-AI052453 and R01 AI0833358 and a Veterans Affairs Merit Award to R.L.G., NIH R01-AR056667 to B.D.Y., the US National Institute of Environmental Health Sciences (NIEHS) Training Grant ES007148 and the NIEHS Center Grant ES005022 supporting J.J.B., and the Department of Veterans Affairs, NIH AR48805 and the Lupus Research Institute to E.L.G.
Footnotes
Note: Supplementary information is available in the online version of the paper.
Author contributions: J.J.B. performed most of the experiments, analyzed results and wrote the manuscript. T.N., J.M., B.M. and A.W.B. assisted with mouse experiments and reviewed the manuscript. C.C.-Z. and B.D.Y. analyzed RNA-Seq results and reviewed the manuscript. E.L.G. and L.M. provided reagents, helped with the design and interpretation of experiments involving U1 RNA and reviewed the manuscript. R.L.G. supervised and designed experiments and wrote and prepared the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. doi: 10.1016/s1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- 2.El Ghissassi F, et al. A review of human carcinogens–part D: radiation. Lancet Oncol. 2009;10:751–752. doi: 10.1016/s1470-2045(09)70213-x. [DOI] [PubMed] [Google Scholar]
- 3.De Fabo EC, Noonan FP. Mechanism of immune suppression by ultraviolet irradiation in vivo I Evidence for the existence of a unique photoreceptor in skin and its role in photoimmunology. J Exp Med. 1983;158:84–98. doi: 10.1084/jem.158.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sontag Y, et al. Cells with UV-specifc DNA damage are present in murine lymph nodes after in vivo UV irradiation. J Invest Dermatol. 1995;104:734–738. doi: 10.1111/1523-1747.ep12606971. [DOI] [PubMed] [Google Scholar]
- 5.Setlow RB. The wavelengths in sunlight effective in producing skin cancer: a theoretical analysis. Proc Natl Acad Sci USA. 1974;71:3363–3366. doi: 10.1073/pnas.71.9.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender K, Gottlicher M, Whiteside S, Rahmsdorf HJ, Herrlich P. Sequential DNA damage-independent and -dependent activation of NF-κB by UV. EMBO J. 1998;17:5170–5181. doi: 10.1093/emboj/17.17.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devary Y, Rosette C, DiDonato JA, Karin M. NF-κB activation by ultraviolet light not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- 8.Walsh LJ. Ultraviolet B irradiation of skin induces mast cell degranulation and release of tumour necrosis factor-α. Immunol Cell Biol. 1995;73:226–233. doi: 10.1038/icb.1995.37. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 11.Akira S, Hirano T, Taga T, Kishimoto T. Biology of multifunctional cytokines: IL 6 and related molecules (IL 1 and TNF) FASEB J. 1990;4:2860–2867. [PubMed] [Google Scholar]
- 12.Vincek V, Kurimoto I, Medema JP, Prieto E, Streilein JW. Tumor necrosis factor α polymorphism correlates with deleterious effects of ultraviolet B light on cutaneous immunity. Cancer Res. 1993;53:728–732. [PubMed] [Google Scholar]
- 13.Vermeer M, Streilein JW. Ultraviolet B light-induced alterations in epidermal Langerhans cells are mediated in part by tumor necrosis factor-α. Photodermatol Photoimmunol Photomed. 1990;7:258–265. [PubMed] [Google Scholar]
- 14.Schwarz A, et al. Ultraviolet-B–induced apoptosis of keratinocytes: evidence for partial involvement of tumor necrosis factor-α in the formation of sunburn cells. J Invest Dermatol. 1995;104:922–927. doi: 10.1111/1523-1747.ep12606202. [DOI] [PubMed] [Google Scholar]
- 15.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 16.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman RW, et al. U1 RNA induces innate immunity signaling. Arthritis Rheum. 2004;50:2891–2896. doi: 10.1002/art.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griswold DE, et al. Activation of the IL-1 gene in UV-irradiated mouse skin: association with infammatory sequelae and pharmacologic intervention. J Invest Dermatol. 1991;97:1019–1023. doi: 10.1111/1523-1747.ep12492422. [DOI] [PubMed] [Google Scholar]
- 19.Kutsch CL, Norris DA, Arend WP. Tumor necrosis factor-α induces interleukin-1 α and interleukin-1 receptor antagonist production by cultured human keratinocytes. J Invest Dermatol. 1993;101:79–85. doi: 10.1111/1523-1747.ep12360119. [DOI] [PubMed] [Google Scholar]
- 20.Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norval M, Simpson TJ, Bardshiri E, Crosby J. Quantifcation of urocanic acid isomers in human stratum corneum. Photodermatol. 1989;6:142–145. [PubMed] [Google Scholar]
- 22.Gilchrest BA, Eller MS, Yaar M. Telomere-mediated effects on melanogenesis and skin aging. J Investig Dermatol Symp Proc. 2009;14:25–31. doi: 10.1038/jidsymp.2009.9. [DOI] [PubMed] [Google Scholar]
- 23.Kosmadaki MG, Gilchrest BA. The role of telomeres in skin aging/photoaging. Micron. 2004;35:155–159. doi: 10.1016/j.micron.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Wei YD, Rannug U, Rannug A. UV-induced CYP1A1 gene expression in human cells is mediated by tryptophan. Chem Biol Interact. 1999;118:127–140. doi: 10.1016/s0009-2797(98)00118-5. [DOI] [PubMed] [Google Scholar]
- 25.Murphy GM, Dowd PM, Hudspith BN, Brostoff J, Greaves MW. Local increase in interleukin-1-like activity following UVB irradiation of human skin in vivo. Photodermatol. 1989;6:268–274. [PubMed] [Google Scholar]
- 26.Clydesdale GJ, Dandie GW, Muller HK. Ultraviolet light induced injury: immunological and infammatory effects. Immunol Cell Biol. 2001;79:547–568. doi: 10.1046/j.1440-1711.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- 27.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3–dependent infammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supp figures