Nuclear Receptors, RXR & the Big Bang (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 27.
Summary
Isolation of genes encoding the receptors for steroids, retinoids, vitamin D and thyroid hormone, and their structural and functional analysis revealed an evolutionarily conserved template for nuclear hormone receptors. This discovery sparked identification of numerous genes encoding related proteins, termed orphan receptors. Characterization of these orphan receptors, and in particular of the retinoid X receptor (RXR), positioned nuclear receptors at the epicenter of the “Big Bang” of molecular endocrinology. This review provides a personal perspective on nuclear receptors and explores their integrated and coordinated signaling networks that are essential for multi-cellular life, highlighting the RXR heterodimer and its associated ligands and transcriptional mechanism.
Introduction
The discovery of nuclear receptors has its historical roots in endocrinology, and the identification of the lipophilic hormones that function as their ligands (Evans, 1988). Steroid and thyroid hormones along with vitamins A and D were elucidated based on their endocrine origin and the physiologic processes that they regulate. Each of these small molecules is chemically distinct and although there was initially no presumption that they might have a shared signaling mechanism, initial biochemical experiments revealed the presence of an intracellular receptor that upon ligand addition activated transcription of tissue-specific sets of target genes (O'Malley, 1971; Yamamoto, 1985).
The idea of a ‘Nuclear Receptor’ that could directly translate simple chemical changes into distinct physiologic effects persisted for several decades, but the fundamental nature this receptor, its means for recognizing specific chemical ligands, its mode of interaction with the genome, and its mechanism for control of gene transcription were all beyond the limits of classic biochemical analysis. Subsequent investigation into the mechanism of hormone action eventually led to the biochemical, molecular, and genetic characterization of the genes encoding the first steroid receptors in the mid-1980s (Mangelsdorf et al., 1995).
The dawn of a superfamily
The isolation of the first complete steroid receptor cDNAs, the glucocorticoid and estrogen receptors, was transformative (Green et al., 1986; Hollenberg et al., 1985) (Figure 1). Comparison of the sequences revealed a conserved evolutionary template, and it also permitted the delineation of the structural and functional features that foreshadowed the emergence of a nuclear receptor superfamily. Each sequence harbors DNA binding, ligand binding, and transactivation domains (Giguere et al., 1986; Green et al., 1986; Miesfeld et al., 1986). Importantly, access to the cDNAs enabled key experiments needed to test protein function, including mutagenesis of the receptor primary structure to assess the importance of specific amino acids and characterization of the nucleotide code within the target gene's promoter that allows gene-specific expression (Green and Chambon, 1987; Umesono and Evans, 1989; Umesono et al., 1988). As a result of this early work, transcriptional regulation by hormone-receptor complexes was shown to be a fundamental process embedded in the circuitry of extracellular signal transduction by lipophilic endocrine hormones and vitamins.
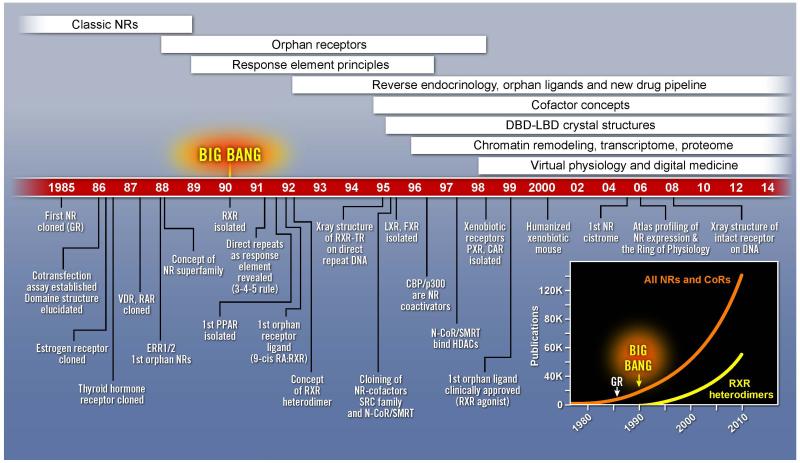
Figure 1. Nuclear Receptor Discovery Timeline.
Schematic timeline showing landmark discoveries in the nuclear receptor field over the last three decades. The entries start from the cloning of the first steroid hormone receptor cDNA to more recent ‘omic’ findings. Inset on the right shows total publications for all nuclear receptors as well as that for RXR and heterodimer partners over time.
Perhaps the most revolutionary finding to come from the cloning of the first steroid receptors was the surprising discovery that dozens of other evolutionarily related proteins exist. As the associated small molecule ligands were unknown, they garnered the name “orphan” receptors (Giguere et al., 1988; Milbrandt, 1988; O'Malley, 1990; Wang et al., 1989). Of further phylogenic significance, these orphan receptors were shown to be conserved throughout metazoan evolution although it should be noted that nuclear receptors are absent in protozoans, fungi and plants (Markov and Laudet, 2011; Owen and Zelent, 2000; Robinson-Rechavi et al., 2004).
Engine of Discovery
That there were orphan receptors immediately suggested the existence of a host of previously unknown signaling pathways regulated by a myriad of undiscovered ligands. The question was how might one go about discovering such ligands? The answer came from a remarkable, innovative technological achievement – the co-transfection assay (Giguere et al., 1986). The idea went like this: If the cDNA encoding the receptor was sufficient to reconstitute a hormone response, then expression plasmids harboring the receptor's cDNA could be co-transfected with a hormone-responsive reporter gene to create a highly defined two-component regulatory switch. With the switch flipped “on” by hormone binding, the resulting powerful transcriptional response allowed rapid analysis of the receptor's DNA and ligand binding domains as well as ligand and target gene specificity. The co- transfection assay was so versatile as a cell-based means to study transcription that it quickly became the mainstay of virtually every molecular biology laboratory as well as a pharmaceutical discovery tool.
The co-transfection assay provided a quantitative and highly efficient tool for screening, and as it was extremely sensitive to small molecule ligands, it became the mainstay of “de-orphaning” efforts. Early results showed that the DNA and ligand binding domains of the receptors function autonomously, which led to the idea that one could swap these domains between receptors, and even other transcription factors, and still retain ligand-dependent transactivation (Giguere et al., 1987; Green and Chambon, 1987; Petkovich et al., 1987). With the cDNAs of the orphan receptors in hand, the cotransfection assay was instantly adapted as an unbiased ligand screening method that could be implemented even when the receptor's DNA binding site (or response element) was not precisely known (Chawla et al., 2001). This “reverse endocrinology” concept allowed for high throughput screens of natural, synthetic and xenobiotic ligands that required less than micromolar concentrations to unearth which receptors might mediate their signaling (Kliewer et al., 1999). In comparison, the classic endocrinology methods that led to the isolation of thyroid hormone in 1914 required 3.5 tons of bovine thyroid gland (Kendall, 1917). Thus, the massive shift in ligand screening sensitivity, as a consequence of receptor cloning and powerful reporter assays, could now unveil new and unimagined signaling systems.
RXR & the Big Bang
Applying the reverse endocrinology approach to the retinoid X receptor (RXR) led to the identification of the first endogenous ligand for an orphan nuclear receptor (9-cis retinoic acid, a metabolite of vitamin A), establishing RXR as the founding member of the “adopted” orphan class (Mangelsdorf et al., 1990; Heyman et al., 1992; Levin et al., 1992) (See Table 1 for receptor nomenclature for the entire superfamily). The discovery of RXR and its ligand resulted in the genesis of two prodigious concepts in the nuclear receptor field. First, in revealing the existence of a previously unknown signaling pathway, it provided a proof of principle that precipitated a wave of research aimed at linking other orphan receptors to specific ligands.
| Common Name | Gene Symbols | Unified Nomenclature | Ligands |
|---|---|---|---|
| Androgen receptor | AR | NR3I4 | androgens |
| Constitutive androstane receptor | CAR | NR1I3 | xenobiotics |
| Chicken ovalbumin upstream promoter-transcription factor alpha | COUP-TFα | NR2F1 | |
| Chicken ovalbumin upstream promoter-transcription factor beta | COUP-TFβ | NR2F2 | |
| Chicken ovalbumin upstream promoter-transcription factor gamma | COUP-TFγ | NR2F6 | |
| Dosage-sensitive sex reversal-adrenal hypoplasia congenital critical region on the X chromosome, gene 1 | DAX-1 | NR0B1 | |
| Estrogen receptor alpha | ERα | NR3A1 | estrogens |
| Estrogen receptor beta | ERβ | NR3A2 | estrogens |
| Estrogen related receptor alpha | ERRα | NR3B1 | |
| Estrogen related receptor beta | ERRβ | NR3B2 | |
| Estrogen related receptor gamma | ERRγ | NR3B3 | |
| Farnesoid X receptor alpha | FXRα | NR1H4 | bile acids |
| Farnesoid X receptor beta* | FXRβ | NR1H5 | |
| Germ cell nuclear factor | GCNF | NR6A1 | |
| Glucocorticoid receptor | GR | NR3CI | glucocorticoids |
| Hepatocyte nuclear factor 4 alpha | HNF4α | NR2A1 | |
| Hepatocyte nuclear factor 4 gamma | HNF4γ | NR2A2 | |
| Liver receptor homolog-1 | LRH-1 | NR5A2 | |
| Liver X receptor alpha | LXRα | NR1H3 | oxysterols |
| Liver X receptor beta | LXRβ | NR1H2 | oxysterols |
| Mineralocorticoid receptor | MR | NR3C2 | mineralocorticoids, glucocorticoids |
| Nerve growth factor induced gene B | NGF1-B | NR4A1 | |
| Neuron-derived orphan receptor 1 | NOR-1 | NR4A3 | |
| Nur-related factor 1 | NURR1 | NR4A2 | |
| Photoreceptor-cell specific nuclear receptor | PNR | NR2E3 | |
| Peroxisome proliferator-activated receptor alpha | PPARα | NR1C1 | fatty acids |
| Peroxisome proliferator-activated receptor beta/delta | PPARβ/δ | NR1C2 | fatty acids |
| Peroxisome proliferator-activated receptor gamma | PPARγ | NR1C3 | fatty acids |
| Progesterone receptor | PR | NR3C3 | progesterone |
| Pregnane X receptor | PXR | NR1I2 | endobiotics, xenobiotics |
| Retinoic acid receptor alpha | RARα | NR1B1 | retinoic acids |
| Retinoic acid receptor beta | RARβ | NR1B2 | retinoic acids |
| Retinoic acid receptor gamma | RARγ | NR1B3 | retinoic acids |
| Reverse-erb alpha | REV-ERBα | NR1D1 | heme |
| Reverse-erb beta | REV-ERBβ | NR1D2 | heme |
| RAR-related orphan receptor alpha | RORα | NR1F1 | |
| RAR-related orphan receptor beta | RORβ | NR1F2 | |
| RAR-related orphan receptor gamma | RORγ | NR1F3 | |
| Retinoid X receptor alpha | RXRα | NR2B1 | 9-cis retinoic acid, docosahexanoic acid |
| Retinoid X receptor beta | RXRβ | NR3B2 | 9-cis retinoic acid, docosahexanoic acid |
| Retinoid X receptor gamma | RXRγ | NR2B3 | 9-cis retinoic acid, docosahexanoic acid |
| Steroidogenic factor 1 | SF-1 | NR5A1 | |
| Short heterodimeric partner | SHP | NR0B2 | |
| Tailless homolog orphan receptor | TLX | NR2E1 | |
| Testicular orphan receptor 2 | TR2 | NR2C1 | |
| Testicular orphan receptor 4 | TR4 | NR2C2 | |
| Thyroid hormone receptor alpha | TRα | NR1A1 | thyroid hormones |
| Thyroid hormone receptor beta | TRβ | NR1A2 | thyroid hormones |
| Vitamin D receptor | VDR | NR1I1 | 1α,25-dihydroxyvitamin D3, lithocholic acid |
Second, it led to the discovery of RXR heterodimerization with these adopted orphan receptors, thereby defining a novel feature of multiple intertwined signaling pathways (Figure 2). For example, one of the first uses of the co- transfection assay identified interactions between an orphan receptor and compounds that promote peroxisome proliferation (fibrates and nafenopin) as the first xenobiotic ligand-receptor pair. The then-named peroxisome proliferator-activated receptor (PPAR) was ultimately shown to be one of a family of fatty acid receptors (Issemann and Green, 1990; Dreyer et al., 1992). PPARs were the first class of orphan receptors shown to form heterodimerize with RXR (Kliewer et al., 1992).
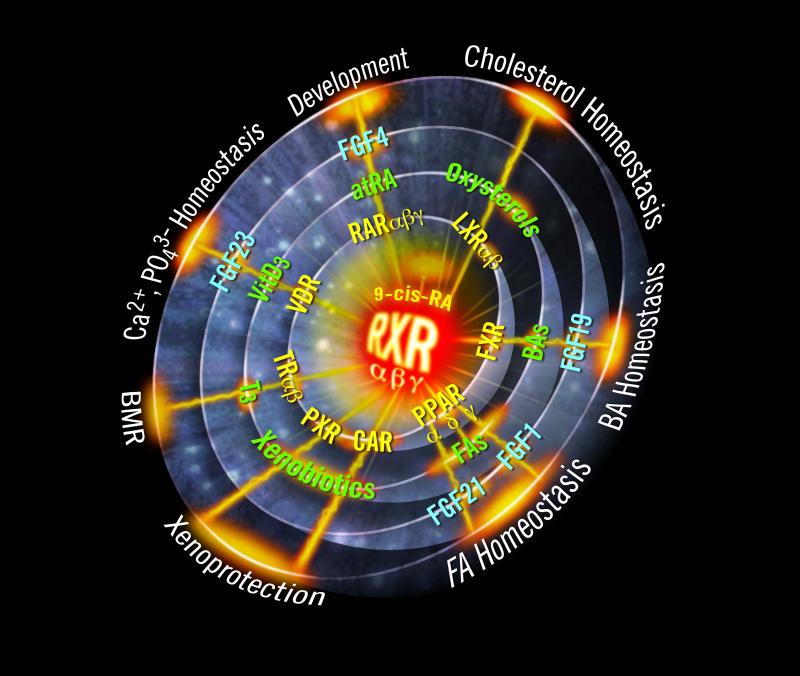
Figure 2. The RXR Big Bang.
The cloning of the RXRs as receptors for 9-cis retinoic acid initiated an expanding wave of discoveries that included: 1) the ability of RXRs to heterodimerize with numerous other nuclear receptors as a mechanism to control gene-specific transcription; 2) the characterization of dietary lipid metabolites (i.e., oxysterols, bile acids, fatty acids, and xenobiotic lipids) as ligands for RXR-partnered orphan receptors; 3) the elucidation of novel endocrine and paracrine signaling pathways mediated by RXR heterodimers, including the fibroblast growth factors 1, 19 and 21; and 4) the role of these receptors in diverse developmental and metabolic pathways. BMR, basal metabolic rate
We refer to the discovery of heterodimerization as the “RXR Big Bang” because its explosive impact gave rise to a wave of discovery, including entirely new physiologic signaling pathways (Figure 2). The straightforward co-transfection assay yielded a relative familiar result – the receptor proteins are functional heterodimers, and yet the complexity of regulation that has since emerged, under both normal and pathological conditions, is nothing short of astonishing.
What's My Ligand?
Classic endocrinology identified thyroid hormone and vitamins A and D as vital factors, however these hormones bind their receptors with high affinity, similar to the classic steroid hormones. In contrast, the reverse endocrinology strategy allowed for the identification of lower-affinity endogenous ligands that were derived from dietary lipids (Kliewer et al., 1999). Furthermore, a major role of the receptors for these lipid-derived ligands was to maintain the homeostasis of the ligands themselves. Thus, the natural ligands being identified turned out to be predictive hallmarks of the physiologic pathways being regulated by their cognate receptors. For example, the finding that fatty acids are endogenous ligands for PPARs led to the discovery that PPARs govern fatty acid metabolism. Likewise the binding of cholesterol metabolites by LXR predicted its future role in controlling cholesterol metabolism. This key bit of fortune, though in retrospect may appear obvious, was decidedly not so at the time. Because the simple chemical structure of the ligand could foretell the intrinsic function of the receptor, ligand discovery became the clarion call of the field. Indeed, physiologic links quickly were established between the growing list of orphan receptors that were being adopted into the nuclear receptor superfamily. These included fatty acid metabolism for the PPARs, sterol homeostasis for the liver X receptors (LXRs), bile acid homeostasis for the farnesoid X receptor (FXR), and endobiotic/xenobiotic metabolism for pregnane X and constitutive androstane receptors (PXR and CAR) (Chawla et al., 2001; O'Malley, 1990). Of further import, these discoveries elaborated the existence of a nuclear receptor superfamily and elucidated the unifying mechanism by which chemically diverse classes of hormones and biologically active lipids coordinately regulate gene expression and diverse physiological pathways.
Make It & Break It
A defining feature of hormone action is the dynamic synthesis and subsequent degradation of receptor ligands. After all, this feature is what drives the recurring nature of the signaling circuits within the physiologic process. Ligand synthesis and its subsequent distribution delivers a body-wide hormonal response via activation of receptor-mediated transcription, while receptor degradation limits both duration and intensity of the response by returning the system to its homeostatic baseline where it's ready for subsequent rounds of activation. For this cyclic process to occur, the ligands themselves must first be produced during the inductive phase and then eliminated to reset the balance. Thus, like the second law of thermodynamics, for every physiologic action, there is an equal, but opposite reaction.
The unsung heroes in this dynamic process are the cytochrome P450s (CYPs), whose unique ability to add molecular oxygen to small molecules plays a key role in cholesterol, steroid and fatty acid metabolism (Baker, 2011; Zelko and Negishi, 2000). For example, all natural steroid hormones (glucocorticoids, mineralocorticoids, progesterone, estrogens and androgens) are synthesized from cholesterol in the gonads and adrenal glands when CYP11A1 (in the mitochondria) converts cholesterol to pregnenolone – the first reaction in steroidogenesis (Strushkevich et al., 2011; Hanukoglu, 1992). A dozen more CYPs are specialized to advance pregnenolone metabolism to the individual hormonal products. This unique relationship between steroid receptors and CYPs (as the ligand generators) extends to virtually all nuclear receptors with endogenous ligands. Distinct CYPs play central roles in production of vitamins A and D for the RARs, RXRs and VDR, as well for synthesis of oxysterols for LXRs, bile acids for FXRs, and fatty acids and arachidonic acid for PPARs (Blumberg and Evans, 1998; Kliewer et al., 1998; McSorley and Daly, 2000; Ray et al., 1997; Russell, 2009).
By virtue of their daily role in hormone synthesis, CYPs also take center stage in receptor ligand inactivation and clearance, a process mostly driven in the liver by the phase I, II and III response, which are controlled in large part by the xenobiotic receptors PXR and CAR. In addition to recognition of foreign compounds (xenobiotics), these receptors also recognize and are activated by endobiotic compounds: steroids, sterols, retinoids, thyroid hormone, and bile acids (Kliewer et al., 1998; Blumberg et al., 1998; Wei et al., 2000; Zelko and Negishi, 2000).
In understanding the unique evolution of nuclear receptors as ligand-dependent transcription factors, it is of interest to note that an important function of nuclear receptors is to autoregulate their own activity by governing the transcription of the specific CYPs that in turn control the concentration of their cognate ligands. Examples of this regulatory circuit include the feedback inhibition of steroid hormone synthesis via the hypothalamic pituitary target organ axis (Dallman et al., 1994; Seminara and Crowley, 2001), bile acid synthesis via the enterohepatic axis (de Aguiar Vallim et al., 2013), and the feedforward regulation of oxysterol, fatty acid, and vitamin D syntheses that are governed by their respective receptors (Chawla et al., 2001; Ory, 2004). Taking this regulatory circuit one step further, the ‘make it and break it’ process uses a special class of orphan nuclear receptors, including SF-1, DAX-1, LRH-1, and SHP, to sequentially produce and eliminate ligands for receptors (PXR and CAR) so that the homeostatic process can be maintained. Furthermore, given the hydrophobic nature of nuclear receptor ligands, ligand transport is coordinately regulated by the cognate receptor, including the production of specific binding proteins and cellular transporters (Pardridge, 1981; Watanabe et al., 1991).
The Heterodimer Rule
As transcription factors, nuclear receptors rely on DNA sequence-specific binding to transactivate their target genes. Utilizing a combination of approaches that included the co-transfection assay and the electrophoretic mobility shift assay, it was determined that the classic steroid receptors (GR, PR, AR, and ER) bind as homodimers to response elements configured as palindromes composed of two hexad nucleotide sequences separated by 3 base pairs (Beato, 1991).
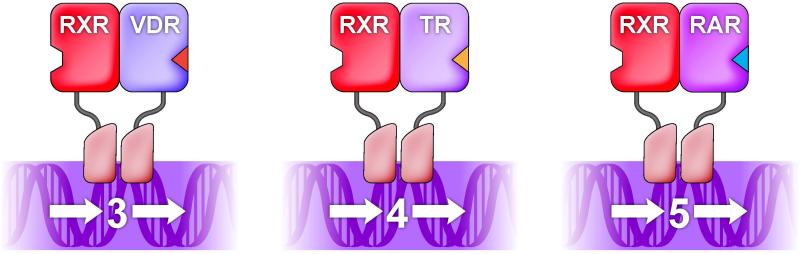
In contrast, non-steroid receptors (i.e., RAR, VDR, TR), bind preferentially to response elements composed of two hexad half-sites arranged as tandem repeats (Koenig et al., 1987; Naar et al., 1991; Umesono et al., 1991). Further, the specificity of each of these receptor/DNA interactions was shown to be encoded uniquely by the nucleotide spacing between the two half-sites of the direct repeat. Thus, the response element for vitamin D was shown to be a direct repeat spaced by 3 nucleotides, for thyroid hormone the spacing is 4 nucleotides, and for retinoic acid it's 5 nucleotides (an axiom that became known as the 3-4-5 rule) (Figure 3) (Perlmann et al., 1993; Umesono et al., 1991). However, perhaps the most striking difference between these receptors and their steroid-binding, homodimeric counterparts was that the non-steroid receptors bind DNA as part of a heterodimer in which their common partner is RXR (Yu et al., 1991; Kliewer et al., 1992; Bugge et al., 1992; Leid et al., 1992; Marks et al., 1992; Zhang et al., 1992). There are now known to be three RXRs (RXRα, RXRβ, and RXRγ), at least one of which is expressed in every cell in the body. The three RXR proteins are highly conserved and functionally interchangeable both as heterodimer partners and as receptors for 9-cis retinoic acid (Mangelsdorf et al., 1992).
Figure 3. The 3-4-5 rule.
Receptors bind DNA as monomers to a single hexad motif, as homodimers to a palindrome of two hexad motifs, or as RXR heterodimers to a tandem repeat of the hexad motif. Heterodimers bind AGGTCA direct repeats (DRs) spaced by 3 (DR3; vitamin D response element), 4 (DR4; thyroid response element) or 5 (DR5; retinoic acid response element) nucleotides as described in the 3-4-5 rule.
The paradigm of RXR heterodimerization quickly expanded into the universe of orphan receptors that now included the PPARs, LXRs, FXR, PXR, and CAR. The success of this paradigm was due in large part because it provided a simple, but elegant solution to the evolution of target gene specificity. On its own, RXR was shown to function as a self-sufficient homodimer, binding to a direct repeat of half-sites separated by 1 nucleotide (i.e., a DR1 element). The DR1 sequence was ideally poised for evolution, because it only required sequential introduction of single nucleotides in the spacer between the two DR1 half-sites to generate novel binding motifs (DR2, DR3, DR4, etc.). A key driver of that process was the innate structure of the RXR ligand binding domain that permits it to adopt multiple conformations and thereby dimerize with different nuclear receptors when bound to each of the direct repeat motifs (Rastinejad et al., 1995; Chandra et al., 2008; Lou et al., 2014). In this way, each new heterodimer partner can bind to one of the half-sites, while RXR can continue to occupy the other because of the flexibility of its dimerization domain (Perlmann et al., 1993). This plasticity allowed the heterodimer rule to expand from the prototypical 3-4-5 rule to include DR1 through DR5 response elements selective for each RXR/receptor partner pair. Although in nature RXR heterodimers have adapted the ability to bind response elements that vary from the canonical direct repeats, the veracity of the general rule has been confirmed by numerous genome-wide binding analyses.
The crystal structures of the heterodimeric unit on DNA have elucidated the molecular basis for these interactions (Rastinejad et al., 1995; Chandra et al., 2008; Lou et al., 2014). The power of the direct repeat rule is that it offered every heterodimer a molecular binding target even in advance of knowledge of its regulatory function. Even so, with 5 different binding sites (6 if we include the single inverted repeat for RXR/FXR), there are still more heterodimers than binding sites. Thus, as would be expected, individual sites can be the target of multiple receptor heterodimers. This promiscuity adds a layer of complexity that may be explained, in part, by tissue specific expression of individual receptors complexes or perhaps features in the chromatin environment that help to ‘craft’ specific binding environments.
Another interesting general property of the RXR heterodimer is that, with only one or two special exceptions, the heterodimer partners are all ligand-dependent. Indeed, a curious evolutionary oddity of the nuclear receptor superfamily has been the rather surprising finding that not all of the orphans have ligands that bind in a reversible, regulatory fashion. For those orphan receptors where such ligands have not been forthcoming, structural, biochemical and physiological evidence indicate that the ligand binding pocket is either absent or occupied continually by a lipid (Wisely et al., 2002), phospholipid (Krylova et al., 2005; Li et al., 2005), or hydrophobic molecule such as heme (Raghuram et al., 2007; Reinking et al., 2005; Yin et al., 2007). Consequently, these orphan receptors generally behave as constitutive activators or repressors of transcription. With respect to their DNA binding, they also function predominantly as monomers or homodimers.
Of the 48 nuclear receptors in humans, approximately half of them fall into the subclass lacking traditional ligands. This observation is of evolutionary interest, as this subclass comprises the most ancient members of the nuclear receptor superfamily, and homologs are found all the way down to the most primitive invertebrate species. Notably, the only invertebrate species where ligand-dependent nuclear receptors have been discovered thus far have been nematodes and arthropods. In C. elegans for example, only one out of 284 nuclear receptors has been characterized as ligand-dependent (Motola et al., 2006; Taubert et al., 2011). In Drosophila, which has 21 nuclear receptors, only two have been confirmed to have ligands (Fahrbach et al., 2012). One of these is the ecdysteroid receptor, and it is of particular interest because it functions as an obligate heterodimer with ultraspiracle, the RXR homolog in insects (Oro et al., 1990; Yao et al., 1992). Thus, the appearance of RXR heterodimerization during evolution has been coincident with the dramatic expansion of ligands for these receptors.
The Silent Partner
An inherent characteristic of the receptor heterodimer is its potential to be activated by either the RXR ligand or the partner receptor ligand. This structural reflection of dual-ligand regulation can be divided into two categories: permissive and non-permissive heterodimers (Forman et al., 1995; Kurokawa et al., 1993). Permissive heterodimers are those that can be activated by ligands of either RXR or its partner, whereas non-permissive heterodimers are those that can only be activated by the partner's ligand while RXR is silent. An important regulatory feature of permissive receptor partners (PPARs, LXRs, FXR, PXR, and CAR) is that the simultaneous presence of both RXR and partner receptor ligands results in a cooperative, synergistic response compared to that resulting from binding of only a single receptor ligand (Leblanc and Stunnenberg, 1995).
Dual-ligand regulation of permissive heterodimers can have both physiologic and pharmacologic consequences. It is of physiologic relevance that all of the permissive receptor partners respond to dietary-derived lipids, whose levels can vary widely. Yet, because of their synergistic relationship with the RXR ligand, even relatively small changes in these lipids can result in robust transcriptional activity and thus achieve a profound biological response. Although endogenous RXR ligands have not been confirmed in every tissue, it is conceivable that the presence of even trace amounts of such ligands would enable a synergistic response. With that in mind, it is of interest to note that in addition to 9-cis retinoic acid, other tissue-specific RXR ligands (e.g., docosahexanoic acid in brain) have been identified (de Urquiza et al., 2000). From a pharmacologic perspective, a number of potent synthetic RXR ligands (called “rexinoids”) have also been described (Mukherjee et al., 1997); and because of their ability to simultaneously activate several heterodimers, such pan-agonists may have utility against multiple therapeutic targets (Desvergne, 2007). While the pharmacologic importance of RXR as a therapeutic target is clear, proof of the physiologic importance of endogenous RXR ligands awaits experiments to effect genetic loss of ligand binding in both individual and combinations of each of the RXR isoforms.
In contrast to the lipid-sensing permissive partners, the non-permissive partners (e.g., TRs, VDR, and RARs) function primarily as hormone receptors and have endocrine ligands that are under tight regulatory control. By silencing RXR activity and responding only to their own ligands, these receptors permit transcriptional regulation that is directly proportional to the level of the hormone, thereby meeting the requirements of endocrine physiology (Shulman and Mangelsdorf, 2005).
The Orphan Receptor Roadmap to Physiology: Engaging the Energy Vector
Homeostasis is essentially a cooperative equilibrium between cells, tissues and organs that relies on a rigorous communication and sensory network as a means to sustain bodily function in all metazoans. In contrast, bacteria and yeast effectively live in their surrounding environment such that every cell must compete for limiting nutrients. In this context, collective cell proliferation usually continues until food sources are depleted and there is no need for a ‘physiologic mechanism’ as all cells are equal.
However in metazoan physiology, cells, tissues and organs serve compartmentalized functions such that the internal environment is distinct from the external environment – thus, the organism must have dynamic, adaptive and co-dependent communication and metabolism at all times. Therefore, real-time control over each metabolic pathway is an essential feature of the homeostatic process. This modulation requires key regulatory sensors to be in place that can monitor levels of specific metabolites and control rate-limiting nodes to reduce or enhance specific routes of energy flux.
The RXR-partnered receptors play a defining role in this process. During the fed state, the body switches on a nuclear receptor network that governs the uptake and storage of energy-rich nutrients, as well as the protection from toxic dietary components (Vacca et al., 2011). For example, in the intestine, FXR is activated by sensing the postprandial increase in bile acids that both facilitate absorption of lipids as well as activate a complex gene expression program to help control diet-linked microbial inflorescence and intestinal inflammation (de Aguiar Vallim et al., 2013). This process serves to create an ‘energy vector’ into the body while maintaining a barrier to the gut microbiome. In part, FXR creates the energy vector by inducing nutrient transporters to bring food into the body and by activating postprandial liver metabolism, through the production of the intestinal hormone FGF19, to help insulin build hepatic glycogen reserves and to recycle and maintain proper bile acid levels (Potthoff et al., 2012).
In concert with this process, activation of LXRs by dietary cholesterol works together with insulin in the liver to promote lipogenesis, further contributing to the energy vector by promoting triglyceride production for delivery to peripheral tissues (Calkin and Tontonoz, 2012). Activation of LXRs in both the gut and the liver also serves to maintain whole body cholesterol homeostasis by up-regulating a gene network that removes excess cholesterol from the body through the process of reverse cholesterol transport and hepatic catabolism of cholesterol into bile acids. To ensure that proper bile acid homeostasis and metabolic balance are maintained, activation of FXR in the intestine and liver activates an important feedback loop to turn off LXR, inhibit further bile acid synthesis, and reset the digestive system once the meal is finished. The second tier of this process is engaged in the periphery by the PPARs to either consume or store excess nutrient lipids. In sensing the rising levels of fatty acids via enhanced energy flux from the liver, PPARα and PPARδ are activated to manage triglyceride and fatty acid metabolism by promoting mitochondrial beta-oxidation and ATP production in muscle and heart (de Lange et al., 2008; Madrazo and Kelly, 2008). The excess energy not consumed is captured and stored in adipose tissue under the control of PPARγ in conjunction with FGF21 and FGF1 to promote adipose remodeling (Dutchak et al., 2012; Jonker et al., 2012; Tontonoz and Spiegelman, 2008). Thus, in a highly cooperative and integrated fashion, the intestine acquires nutrients, the liver transforms and delivers nutrients, and active tissues burn nutrients, whereas adipose collects unused energy for long-term storage without being a major consumer – effectively representing an altruistic function.
While the FXR/LXR/PPAR axis is on the front end of nutrient acquisition, the xenobiotic receptors PXR and CAR are part of the clearance system that allows the body to rid itself of toxic dietary metabolites, drugs and endobiotics (such as steroid, retinoid, and thyroid hormones) (Willson and Kliewer, 2002). The ability of these two receptors to be activated by 1000s of compounds enables the transcription of a detoxification gene program that creates a clearance vector through which toxic body fluids are eliminated through the Phase I, II and III arms of the xenobiotic response.
During privation when nutrients are limiting or absent, the entire energy vector is reversed, and the body now mobilizes its stored energy reserves in adipose to use as fuel while at the same time suppressing unnecessary energy consuming pathways. PPARα is a primary regulator of this adaptive response to starvation, where in the liver it senses the reversed flux of fatty acids and activates a gene network to convert fatty acids into a usable energy source (i.e., ketone bodies) (Contreras et al., 2013). Activation of PPARα also results in the production of the hepatokine, FGF21, which sends a stress signal to other sites in the body thereby facilitating the body's adaptation to an energy-deprived state (Potthoff et al., 2012).
Thus, unlike simple organisms where the energy demands of each member of the population are approximately equal, in metazoans such demands are highly variable. As direct sensors of dietary lipids, bile acids and their metabolites, RXR-partnered receptors are key in controlling the dynamics of energy flow and organ communication. This is summarized in Figure 4.
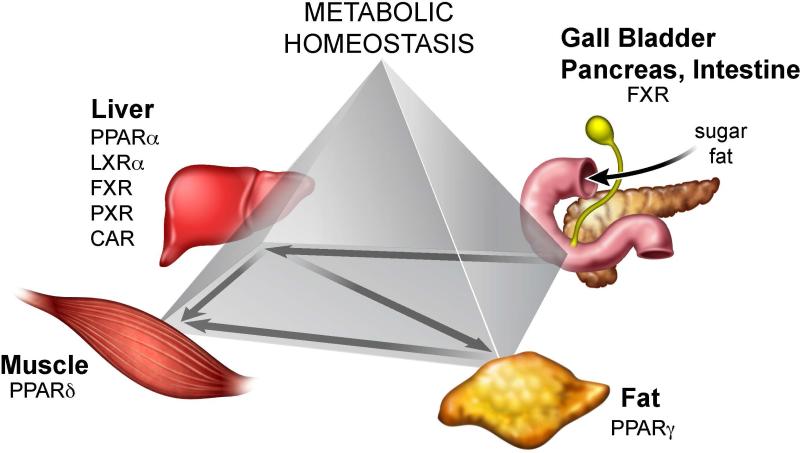
Fig 4. Metabolic homeostasis & the energy vector.
The figure illustrates that nutrients, such as sugar and fat, enter the body and are processed in a highly vectorial fashion. Vector 1: The gallbladder, pancreas and intestine are the most important digestive organs in the body. Detergent properties of bile acids both solubilize lipids to promote lipid absorption and by activating FXR to transiently induce 100s of genes for nutrient transport and suppression of microbial activity along with the release of FGF19 as a hormonal signal to the liver. In the liver, PPARα is activated to break down fatty acids and induce FGF21 as a hormonal signal to adipose (and other tissues in the body). LXR induces Cyp7a1 to convert cholesterol into bile acid and SREBP-1c activation for fatty acid synthesis. Vector 2: PPARδ activation during exercise triggers production of sugar and fat in the liver for delivery to muscle for both glycolytic and oxidative metabolism. Vector 3: unconsumed energy is sent to fat where under the control of the PPARγ-FGF1 and -FGF21 axis, nutrients are stored in adipose tissue. Vector 4: In response to demand, nutrients stored in visceral fat are sent to muscle or other tissues.
Novel therapeutics on the ‘event horizon’
An immediate implication that followed from the initial discovery of the receptors for the steroid and thyroid hormones, and vitamins A and D, was their potential as therapeutic targets. Indeed, drugs that target these receptors are amongst the most widely used and commercially successful. Thus, it should not be surprising that the RXRs and their–orphan receptor partners might become the next generation of relevant therapeutic targets. Indeed, bexarotene and alitretinoin (RXRs), fibrates (PPARα), and thiazolidinediones (PPARγ) are already approved drugs for treating cancer, hyperlipidemia, and type 2 diabetes, respectively (Moore et al., 2006). Looking out on the ‘event horizon’ of drug discovery, it is notable that FXR and LXR agonists are in development for treating nonalcoholic steatohepatitis and preventing atherosclerosis. Perhaps just as importantly, PXR is now used routinely in the pharmaceutical industry to screen all new drug candidates for potentially dangerous drug-drug interactions.
Beyond the Big Bang
It is worth mentioning that another consequence of the RXR Big Bang has been the discovery of several other orphan receptors that integrate into the Ring of Physiology (Bookout et al., 2006; Yang et al., 2006). The Nuclear Receptor Ring of Physiology is a circular dendogram based on the unsupervised clustering of nuclear receptor mRNA expression in 39 mouse tissues that revealed the relationship between nuclear receptor expression, function and physiology (Bookout et al., 2006). Many of these receptors, such as the RORs and Rev-Erbs that govern circadian clocks (Guillaumond et al., 2005) and the ERRs that govern energy homeostasis (Giguere, 2008), are notable for their coordinate regulation of physiologic networks that often intersect with the RXR heterodimers.
Genomic gymnastics – dynamic physiology from dynamic genomes
An unmet future goal of the nuclear receptor field, and indeed of transcription factors in general, is understanding how they govern the transcriptional process. As ligand-dependent transcription factors, nuclear receptors like the RXR heterodimers must be understood as a consequence of their ability to interact with the genome and promote chromatin remodeling that, via epigenetic changes, in turn modulate gene expression. This process must be dynamic, affecting elaborate networks of genes and global patterns of gene expression. As this regulation is hormone-dependent, induction of this regulatory triad (DNA binding, chromatin modulation and transcriptional activation) must be reversed when ligand levels recede. In essence, physiologic complexity must arise from genomic complexity reflected by a coordinated genome-wide interaction of the receptors with DNA. One key to understanding this physiologic complexity has come from the discovery of “pioneering factors” that function as tissue-specific transcriptional programmers. By helping to guide cell lineage, pioneer factors set the genomic landscape through which signal dependent transcription factors can activate target gene networks. Several studies have pointed to the role pioneer factors play in collaborating with nuclear receptors in a cell-type specific manner (Heinz et al., 2010). In this way, the same nuclear receptor is able to govern distinct transcriptional networks in different cell types (Miranda et al., 2013).
Cistromes
In the past 20 years, we have learned that a major function of RXR heterodimers is to act as a reversible switch, which in the absence of ligand recruits a complex of factors (i.e, co-repressors) to target gene promoters to repress transcriptional output, whereas in the presence of the ligand the heterodimer recruits co-activator complexes to open up chromatin and turn up transcription of target genes (O'Malley et al., 2012; Rosenfeld et al., 2006). This view, while still largely correct, is only one layer of the complexity of the process. To understand the complete logic of how these receptors function, one needs to transition from probing single DNA binding elements near the promoter to surveying chromatin interactions at the genome-wide scale. The development of high-throughput techniques to analyze these receptor-DNA interactions (e.g., ChIP-seq) has not only confirmed the structure-function rules for nuclear receptor response elements, but it has also led to a number of unexpected results. A rather surprising finding is that the sum total of individual binding sites (cumulatively called a cistrome) for a single receptor in a typical cell may comprise 10,000-25,000 sites within 250-1000 genes (Tang et al., 2011). What do these many thousands of binding events tell us about hormone signaling? Are they all important and if not, is there a way to decipher those that are functional from those that are inactive decoys? Minimally, cistromic analysis is valuable as it allows the positioning of RXR-heterodimers in relation to specific genes. Unlike steroid receptors that enter the nucleus with ligand treatment, RXR heterodimers remain nuclear (and bound to many of these sites) whether activated by ligand or not. In this context, RXR heterodimers are unusual in the sense that a single bound factor at a single site can cycle as both a positive and negative regulator, depending on the presence or absence of a ligand. An interesting general feature from cistromes is that motif analysis can only confirm binding to known sites. Thus, divergent DNA binding elements may be present, but difficult to identify. In addition, as at least 1/3 or more binding events have no clear hormone response element, perhaps this fraction of chromatin bound NRs are present due to direct recruitment to other transcription factors (tethering).
While cistromic analysis can uncover multiple cooperative, interactive and synergistic relationships it still represents only a ‘snapshot’ of a very dynamic and possibly short-lived, process. Though fragmentary, significant evidence is accumulating that in any one cell type, cistromes can expand and contract depending on developmental or environmental signals that trigger expression or influx of transcription factors such as FOXa1 NF-κB, SMADs & STATs into the nucleus. This process clearly can expose large numbers of cryptic nuclear receptor binding elements. As just one example, in hepatic stellate cells RXR/VDR heterodimers bind 6,281 sites in quiescent cells and 24,984 sites following LPS and TGFβ-activation (Ding et al., 2013). What triggers movement of RXR/VDR to new sites is not known, but seems to be important as it allows nuclear receptors to be recruited to regions of genomic activity, even if these are only transient in nature. The ability to move ensures that hormone-responsive gene regulation can participate in the control of all key gene networks in real-time as these networks are themselves being formed. Thus, the mechanism of “motional adaptability” is important to explore as it underpins the rapid and dynamic features of hormone signaling.
Enhancers and Transcriptomes
While cistromes can be dynamic, they are still a type of ‘genographical’ homing mechanism that needs to be placed in context of the enhancer. Enhancers represent the clustering of cis-regulatory elements that can confer both positive and negative regulation on target genes. In general, the average distance of an enhancer from a promoter is ~100kb, greatly strengthening the idea that the enhancer functions, in part, via DNA looping with the regulated promoter (Sanyal et al., 2012). Whether loops themselves are hormone-inducible has not been established and whether hormone repression involves loop disruption needs to be addressed.
New techniques such as global run-on sequencing (GRO-Seq) can measure near instantaneous changes in expression in a sensitive and quantitative fashion. GRO-Seq analysis unexpectedly revealed that upstream enhancers produce short non-coding transcripts known as eRNAs (Hah and Kraus, 2014). Recent work (Lam et al., 2013) supports the idea that orphan nuclear receptors can dial up or down the transcriptional output of the eRNA in a fashion linked to the transcriptional activity of the downstream gene itself (Hah et al., 2011). While this is still very early days, the idea that eRNAs are ligand-responsive leads us to wonder whether enhancer transcripts represent the initial level of hormonal control of gene expression. Addressing this important question will undoubtedly comprise a major feature of future analysis.
One other class of specialty components of the transcriptome that deserves mention are the microRNAs, which by virtue of hybridizing with a complementary sequence, repress gene expression. Their relationship to RXR-heterodimer signaling is just now coming to light. For example, in the case of muscle thyroid hormone signaling, expression of a single miRNA represses type I myofiber formation to produce a fast muscle phenotype. Thus, via their receptor-mediated expression, miRNAs can silence large networks of genes and thereby play a key role in mediating the hormonal response (Gan et al., 2013; van Rooij et al., 2007; Williams et al., 2009). Future studies are needed to catalogue and characterize nuclear receptor-regulated miRNAs and their contribution to both tissue and systemic physiology.
Although genome-wide DNA sequencing technology helped to create a map of coding sequences, introns and regulatory sites, sequence information by itself is static. What we are learning is that although there is only one genome in a person, there will be many “epigenomes” throughout the body, and that even in small clusters of cells such epigenomes may be dynamic. This epigenetic interface is now where a deeper understanding of nuclear receptor-regulated complex physiologic processes will begin to emerge.
Expanding the NR Universe
Progress in molecular biology, genetics, chemistry and structure has led to a vast accumulation of knowledge about the identities, regulation and function of RXR and its heterodimeric partners since their original discovery over 25 years ago. Much of this progress has been facilitated by striking advances in physiological techniques and model in vivo systems, as well as in ‘omic’ technologies. As these technologies have continued to advance, so have the critical informatic methods to interrogate and compare the massive data being generated. Yet, although much has been learned about each of the adopted orphan RXR partners, it seems we have only begun to scratch the surface on a true understanding of their physiologic functions. The challenge represents a computational window into the complexity of the physiologic process itself.
Conclusions
By taking a step back, in this perspective, we have tried to provide a broad overview of the cumulative progress in understanding nuclear receptor function in classic endocrinology, and more importantly to chart the expanding impact of the RXR ‘Big Bang’ on new biology. Physiology reflects integrated body- wide processes with receptors, metabolites, ligand and conjoint signaling molecules in a continual orchestration, much of which is involved in the ‘trafficking’ of nutrients throughout the body, along with hormonal signals that open and close portals to tissues, organs and cells. While many of these events are traceable in blood, a more complete understanding will require a much deeper insight into the brain as a specialized endocrine organ, and how it oversees what we now call physiologic integration.
Nevertheless, great advances have been made and remarkable insights gleaned into dozens of new physiologic pathways as well as into pathology and treatment through modulation of nuclear receptor function. Orphan nuclear receptors have now transitioned through the tipping point as therapeutic targets in the treatment of human disease. Thus, the ‘Big Bang’ is still expanding, enhancing our insight into a modern definition of physiology with unlimited potential to uncover the secrets of the human condition.
Acknowledgements
We thank R. Yu, M. Downes, A. Atkins, S. Kliewer, and N. McKenna for discussions and critical input, J. Simon for artwork, and C. Brondos and E. Ong for administrative assistance. R.M.E. holds the March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute and is supported by grants from NIH (DK057978, HL105278, DK090962, HL088093, ES010337 and CA014195) as well as the Helmsley Charitable Trust, Samuel Waxman Cancer Research Foundation and Ipsen/Biomeasure. D.J.M. is supported by the NIH (R01DK067158) and the Robert A. Welch Foundation (grant I-1275). R.M.E and D.J.M. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker ME. Origin and diversification of steroids: co-evolution of enzymes and nuclear receptors. Mol Cell Endocrinol. 2011;334:14–20. doi: 10.1016/j.mce.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Beato M. Transcriptional control by nuclear receptors. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1991;5:2044–2051. doi: 10.1096/fasebj.5.7.2010057. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Evans RM. Orphan nuclear receptors--new ligands and new possibilities. Genes & development. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W, Jr., Juguilon H, Bolado J, Jr., van Meter CM, Ong ES, Evans RM. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes & development. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge TH, Pohl J, Lonnoy O, Stunnenberg HG. RXR alpha, a promiscuous partner of retinoic acid and thyroid hormone receptors. The EMBO journal. 1992;11:1409–1418. doi: 10.1002/j.1460-2075.1992.tb05186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nature reviews Molecular cell biology. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Contreras AV, Torres N, Tovar AR. PPAR-alpha as a key nutritional and environmental sensor for metabolic adaptation. Advances in nutrition. 2013;4:439–452. doi: 10.3945/an.113.003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell metabolism. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman MF, Akana SF, Levin N, Walker CD, Bradbury MJ, Suemaru S, Scribner KS. Ann N Y Acad Sci. 1994;746:22–31. doi: 10.1111/j.1749-6632.1994.tb39206.x. [DOI] [PubMed] [Google Scholar]
- de Lange P, Lombardi A, Silvestri E, Goglia F, Lanni A, Moreno M. Peroxisome Proliferator-Activated Receptor Delta: A Conserved Director of Lipid Homeostasis through Regulation of the Oxidative Capacity of Muscle. PPAR research. 2008;2008:172676. doi: 10.1155/2008/172676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Urquiza AM, Liu S, Sjoberg M, Zetterstrom RH, Griffiths W, Sjovall J, Perlmann T. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- Desvergne B. RXR: from partnership to leadership in metabolic regulations. Vitamins and hormones. 2007;75:1–32. doi: 10.1016/S0083-6729(06)75001-4. [DOI] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- Dutchak PA, Katafuchi T, Bookout AL, Choi JH, Yu RT, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor-21 regulates PPARgamma activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148:556–567. doi: 10.1016/j.cell.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE, Smagghe G, Velarde RA. Insect nuclear receptors. Annual review of entomology. 2012;57:83–106. doi: 10.1146/annurev-ento-120710-100607. [DOI] [PubMed] [Google Scholar]
- Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- Gan Z, Rumsey J, Hazen BC, Lai L, Leone TC, Vega RB, Xie H, Conley KE, Auwerx J, Smith SR, et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. The Journal of clinical investigation. 2013;123:2564–2575. doi: 10.1172/JCI67652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V. Transcriptional control of energy homeostasis by the estrogen- related receptors. Endocrine reviews. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Giguere V, Hollenberg SM, Rosenfeld MG, Evans RM. Functional domains of the human glucocorticoid receptor. Cell. 1986;46:645–652. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- Green S, Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987;325:75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. Journal of biological rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Hah N, Kraus WL. Hormone-regulated transcriptomes: lessons learned from estrogen signaling pathways in breast cancer cells. Mol Cell Endocrinol. 2014;382:652–664. doi: 10.1016/j.mce.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. The Journal of steroid biochemistry and molecular biology. 1992;43:779–804. doi: 10.1016/0960-0760(92)90307-5. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Jonker JW, Suh JM, Atkins AR, Ahmadian M, Li P, Whyte J, He M, Juguilon H, Yin YQ, Phillips CT, et al. A PPARgamma-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485:391–394. doi: 10.1038/nature10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall EC. The crystalline compound containing iodine which occurs in the thyroid. Endocrinology. 1917;1:153–169. [Google Scholar]
- Kliewer SA, Lehmann JM, Willson TM. Orphan nuclear receptors: shifting endocrinology into reverse. Science. 1999;284:757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig RJ, Brent GA, Warne RL, Larsen PR, Moore DD. Thyroid hormone receptor binds to a site in the rat growth hormone promoter required for induction by thyroid hormone. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:5670–5674. doi: 10.1073/pnas.84.16.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Yu VC, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld MG, Glass CK. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes & development. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc BP, Stunnenberg HG. 9-cis retinoic acid signaling: changing partners causes some excitement. Genes & development. 1995;9:1811–1816. doi: 10.1101/gad.9.15.1811. [DOI] [PubMed] [Google Scholar]
- Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, Zacharewski T, Chen JY, Staub A, Garnier JM, Mader S, et al. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature. 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Molecular cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lou X, Toresson G, Benod C, Suh JH, Philips KJ, Webb P, Gustafsson JA. Structure of the retinoid X receptor α-liver X receptor β (RXRα-LXRβ) heterodimer on DNA. Nat Struct Mol Biol. 2014 2014 Mar;21(3):277–81. doi: 10.1038/nsmb.2778. doi: 10.1038/nsmb.2778. [DOI] [PubMed] [Google Scholar]
- Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. Journal of molecular and cellular cardiology. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, Oro AE, Kakizuka A, Evans RM. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes & development. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov GV, Laudet V. Origin and evolution of the ligand-binding ability of nuclear receptors. Mol Cell Endocrinol. 2011;334:21–30. doi: 10.1016/j.mce.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Marks MS, Hallenbeck PL, Nagata T, Segars JH, Appella E, Nikodem VM, Ozato K. H-2RIIBP (RXR beta) heterodimerization provides a mechanism for combinatorial diversity in the regulation of retinoic acid and thyroid hormone responsive genes. The EMBO journal. 1992;11:1419–1435. doi: 10.1002/j.1460-2075.1992.tb05187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSorley LC, Daly AK. Identification of human cytochrome P450 isoforms that contribute to all-trans-retinoic acid 4-hydroxylation. Biochemical pharmacology. 2000;60:517–526. doi: 10.1016/s0006-2952(00)00356-7. [DOI] [PubMed] [Google Scholar]
- Miesfeld R, Rusconi S, Godowski PJ, Maler BA, Okret S, Wikstrom AC, Gustafsson JA, Yamamoto KR. Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell. 1986;46:389–399. doi: 10.1016/0092-8674(86)90659-8. [DOI] [PubMed] [Google Scholar]
- Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endocrinol. 2013;380:16–24. doi: 10.1016/j.mce.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. ChemMedChem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, Mangelsdorf DJ. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Mukherjee R, Davies PJ, Crombie DL, Bischoff ED, Cesario RM, Jow L, Hamann LG, Boehm MF, Mondon CE, Nadzan AM, et al. Sensitization of diabetic and obese mice to insulin by retinoid X receptor agonists. Nature. 1997;386:407–410. doi: 10.1038/386407a0. [DOI] [PubMed] [Google Scholar]
- Naar AM, Boutin JM, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- O'Malley BW. Mechanisms fo action of steroid hormones. N Engl J Med. 1971;284:370–377. doi: 10.1056/NEJM197102182840710. [DOI] [PubMed] [Google Scholar]
- O'Malley B. The steroid receptor superfamily: more excitement predicted for the future. Mol Endocrinol. 1990;4:363–369. doi: 10.1210/mend-4-3-363. [DOI] [PubMed] [Google Scholar]
- O'Malley BW, Malovannaya A, Qin J. Minireview: nuclear receptor and coregulator proteomics--2012 and beyond. Mol Endocrinol. 2012;26:1646–1650. doi: 10.1210/me.2012-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oro AE, McKeown M, Evans RM. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature. 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- Ory DS. Nuclear receptor signaling in the control of cholesterol homeostasis: have the orphans found a home? Circulation research. 2004;95:660–670. doi: 10.1161/01.RES.0000143422.83209.be. [DOI] [PubMed] [Google Scholar]
- Owen GI, Zelent A. Origins and evolutionary diversification of the nuclear receptor superfamily. Cellular and molecular life sciences : CMLS. 2000;57:809–827. doi: 10.1007/s000180050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge WM. Transport of protein-bound hormones into tissues in vivo. Endocrine reviews. 1981;2:103–123. doi: 10.1210/edrv-2-1-103. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Rangarajan PN, Umesono K, Evans RM. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes & development. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- Petkovich M, Brand NJ, Krust A, Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15 /19 and 21: from feast to famine. Genes Dev. 2012;26(4):312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nature structural & molecular biology. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- Ray WJ, Bain G, Yao M, Gottlieb DI. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. The Journal of biological chemistry. 1997;272:18702–18708. doi: 10.1074/jbc.272.30.18702. [DOI] [PubMed] [Google Scholar]
- Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Boussau B, Laudet V. Phylogenetic dating and characterization of gene duplications in vertebrates: the cartilaginous fish reference. Molecular biology and evolution. 2004;21:580–586. doi: 10.1093/molbev/msh046. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & development. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50:S120–S125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminara SB, Crowley WF., Jr. Perspective: the importance of genetic defects in humans in elucidating the complexities of the hypothalamic-pituitary-gonadal axis. Endocrinology. 2001;142:2173–2177. doi: 10.1210/endo.142.6.8261. [DOI] [PubMed] [Google Scholar]
- Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. The New England journal of medicine. 2005;353:604–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- Strushkevich N, MacKenzie F, Cherkesova T, Grabovec I, Usanov S, Park HW. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10139–10143. doi: 10.1073/pnas.1019441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Chen Y, Meyer C, Geistlinger T, Lupien M, Wang Q, Liu T, Zhang Y, Brown M, Liu XS. A comprehensive view of nuclear receptor cancer cistromes. Cancer research. 2011;71:6940–6947. doi: 10.1158/0008-5472.CAN-11-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Ward JD, Yamamoto KR. Nuclear hormone receptors in nematodes: evolution and function. Mol Cell Endocrinol. 2011;334:49–55. doi: 10.1016/j.mce.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annual review of biochemistry. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Umesono K, Evans RM. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Umesono K, Giguere V, Glass CK, Rosenfeld MG, Evans RM. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca M, Degirolamo C, Mariani-Costantini R, Palasciano G, Moschetta A. Lipid-sensing nuclear receptors in the pathophysiology and treatment of the metabolic syndrome. Wiley Interdiscip Rev Syst Biol Med. 2011;3:562–587. doi: 10.1002/wsbm.137. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wang LH, Tsai SY, Cook RG, Beattie WG, Tsai MJ, O'Malley BW. COUP transcription factor is a member of the steroid receptor superfamily. Nature. 1989;340:163–166. doi: 10.1038/340163a0. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Tani T, Watanabe S, Seno M. Transport of steroid hormones facilitated by serum proteins. Biochimica et biophysica acta. 1991;1073:275–284. doi: 10.1016/0304-4165(91)90132-z. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Current opinion in cell biology. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nature reviews Drug discovery. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr., Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–52. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yao TP, Segraves WA, Oro AE, McKeown M, Evans RM. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erb alpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Naar AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- Zelko I, Negishi M. Phenobarbital-elicited activation of nuclear receptor CAR in induction of cytochrome P450 genes. Biochem Biophys Res Commun. 2000;277:1–6. doi: 10.1006/bbrc.2000.3557. [DOI] [PubMed] [Google Scholar]
- Zhang XK, Lehmann J, Hoffmann B, Dawson MI, Cameron J, Graupner G, Hermann T, Tran P, Pfahl M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature. 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]