Spotted Long Oligonucleotide Arrays for Human Gene Expression Analysis (original) (raw)
Abstract
DNA microarrays produced by deposition (or `spotting')of a single long oligonucleotide probe for each gene may be an attractive alternative to other types of arrays. We produced spotted oligonucleotide arrays using two large collections of ∼70-mer probes, and used these arrays to analyze gene expression in two dissimilar human RNA samples. These samples were also analyzed using arrays produced by in situ synthesis of sets of multiple short (25-mer)oligonucleotides for each gene (Affymetrix GeneChips). We compared expression measurements for 7344 genes that were represented in both long oligonucleotide probe collections and the in situ-synthesized 25-mer arrays. We found strong correlations (r = 0.8–0.9)between relative gene expression measurements made with spotted long oligonucleotide probes and in situ-synthesized 25-mer probe sets. Spotted long oligonucleotide arrays were suitable for use with both unamplified cDNA and amplified RNA targets, and are a cost-effective alternative for many functional genomics applications. Most previously reported evaluations of microarray technologies have focused on expression measurements made on a relatively small number of genes. The approach described here involves far more gene expression measurements and provides a useful method for comparing existing and emerging techniques for genome-scale expression analysis.
Microarrays can be produced by deposition (or spotting) of DNA or by in situ synthesis of oligonucleotides on a solid substrate. Spotted cDNA arrays are typically produced by depositing PCR amplicons, made from cDNA clones, on modified glass slides (Schena et al. 1996). In general, PCR amplicons are several hundred to a few thousand base pairs, and one amplicon (or sometimes a few different amplicons) are used to probe each gene. These arrays can be produced by individual investigators or core facilities, or can be purchased commercially. Production of microarrays by in situ synthesis requires more sophisticated and costly equipment, and these arrays are generally produced commercially. One widely used implementation of this technology is the Affymetrix short oligonucleotide array (GeneChip). Here, photolithography and solid-phase chemistry are used to produce high-density arrays of 25-mer oligonucleotides (Lockhart et al. 1996). Each perfect-match oligonucleotide is paired with a mismatched oligonucleotide, and several (11–20) pairs of 25-mers are used for each gene.
Various approaches have been used to verify the accuracy of microarray data. Microarray assay technology can be calibrated by spiking known quantities of one or several RNA transcripts into test samples. Alternatively, independent methods including Northern blotting or quantitative PCR can be used to verify array measurements (Yuen et al. 2002). Results from both approaches indicate that spotted cDNA arrays and short oligonucleotide arrays accurately quantify gene expression in many cases. However, some exceptions have been noted, and these approaches typically only evaluate measurements of a very small fraction of the thousands of genes represented on modern microarrays. Other investigators have compared the performance of spotted cDNA arrays and Affymetrix 25-mer arrays across a larger set of genes. Kuo et al. (2002) reported that measurements of gene expression by spotted cDNA arrays and Affymetrix 25-mer arrays showed little correlation. However, this comparison involved data generated by two different groups using RNA samples generated independently from the same set of cell lines, and therefore it is not clear whether the lack of correlation is attributable to the difference in array type. Li et al. (2002) used both commercial cDNA arrays and Affymetrix GeneChips to analyze gene expression changes induced by _tert-_butylhydroxyquinone treatment of human neuroblastoma cells. They concluded that there were very substantial discrepancies between the two array types, and that the cDNA arrays often failed to detect differences in gene expression. Kothapalli et al. (2002) also used both of these array types to analyze gene expression in peripheral blood mononuclear cells from normal and leukemic subjects, and found substantial discrepancies. These discrepancies have been attributed to several factors, including nonspecific hybridization and misidentification of cDNA probes. Thus, the small number of reports that directly address this issue do not provide clear support for the idea that there is good agreement between gene expression measurements made with spotted cDNA arrays and in situ-synthesized short oligonucleotide arrays.
Spotted long oligonucleotide arrays were recently introduced as an alternative to spotted cDNA arrays and in situ-synthesized oligonucleotide arrays (Kane et al. 2000). Spotted oligonucleotide arrays are produced by deposition (or `spotting') of solutions containing synthetic oligonucleotides, typically 40–90 bases long, on a solid substrate. These arrays can be produced and used with methods identical or very similar to those used for spotted cDNA arrays. Although large sets of oligonucleotide probes and arrays made from long oligonucleotide probes are now available from various suppliers, there are as yet few published reports involving the evaluation or application of these arrays. However, strong support for the utility of long oligonucleotide probes comes from a study of 60-mer oligonucleotide arrays fabricated by an ink-jet oligonucleotide synthesizer (Hughes et al. 2001). Results obtained with this type of array correlated well with results obtained using yeast cDNA arrays, and human ink-jet 60-mer arrays were able to identify 20 of 23 human genes that had been found to be upregulated by at least threefold following retinoic acid-induced differentiation of NB4 cells, in two previous studies performed using cDNA arrays and Affymetrix GeneChips.
To evaluate the performance of spotted oligonucleotide arrays, we used these arrays to compare gene expression in two dissimilar RNA samples. We expected that this would result in a large number of differentially expressed genes, which would allow us to draw meaningful conclusions about how well measurements made using different array types were correlated across a large set of genes. We produced arrays using two large commercially available collections of ∼70-mer oligonucleotide probes for thousands of genes (generally one probe per gene). We analyzed gene expression in the two RNA samples using these two types of spotted long oligonucleotide arrays as well as Affymetrix 25-mer arrays. We were able to compare measurements for 7344 genes represented on all three arrays, and found strong correlations between relative expression measurements made using these three different array types.
RESULTS
We produced two different sets of spotted arrays using two collections of long oligonucleotide probes (Operon Human Genome Oligo Set Versions 1 and 2,Table 1). There were 10,801 UniGene clusters that were represented in both groups of probes, but the sequences of these two groups of probes were largely independent: Version 1 and Version 2 probes overlapped significantly (by at least 25 identical bases) for just 1935 of the 10,801 gene clusters that were represented in both versions. We also used commercially produced arrays containing sets of 25-mer probes synthesized in situ (Affymetrix U95Av2 GeneChips). We used all three groups of probes to compare gene expression in two total RNA samples, one made from K562 erythroleukemia cells and one made from a pool of 10 different cell lines.
Table 1.
Probe Characteristics
| Probe group | Compositiona | Distinct GenBank identifiersb | Distinct UniGene clustersc | Not associated with a UniGene clusterc |
|---|---|---|---|---|
| Operon Human Genome Oligo Set Version 1 | 13,971 long oligonucleotide probesd | 13,652 | 11,158 | 2120 |
| Operon Human Genome Oligo Set Version 2 | 21,329 long oligonucleotide probesd | 21,306 | 19,888 | 728 |
| Affymetrix U95Av2 GeneChip | 12,558 sets of oligonucleotides, ∼16 perfect match 25-mers per set | 11,282 | 8328 | 1695 |
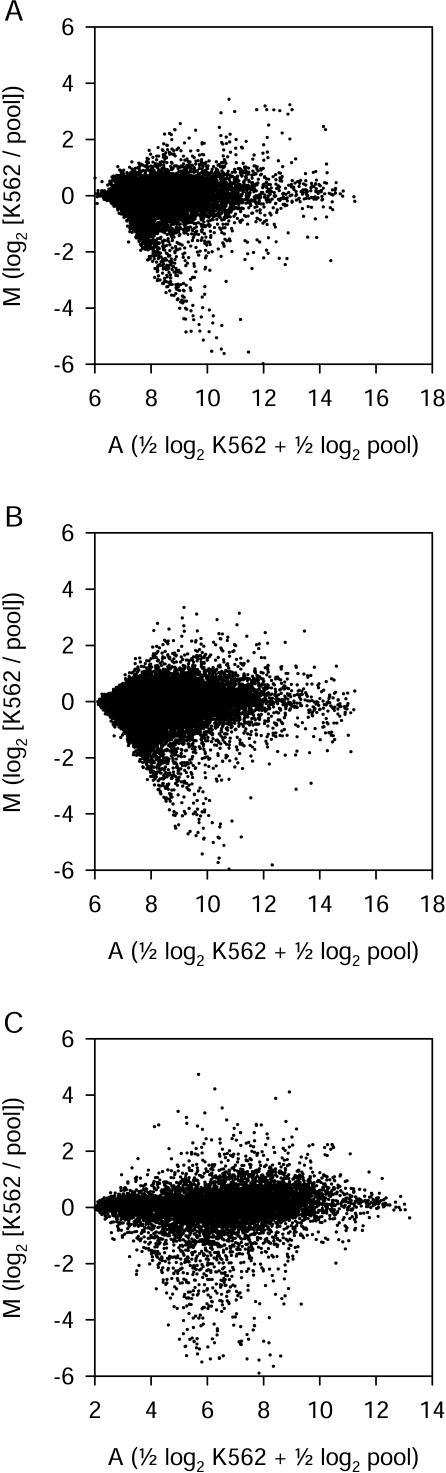
For spotted long oligonucleotide arrays, the RNA samples were used to produce labeled cDNA targets. Two color hybridizations were performed using Cy3- and Cy5-labeled targets derived from the two RNA samples, and gene expression ratios were calculated for each hybridization. Six independent replicates were performed for Version 1 probe hybridizations, and four replicates were performed for Version 2 probe hybridizations. Two values,M and A, were calculated for each element on each array.M is a normalized, log2-transformed measure of differential gene expression. Positive M values indicate higher normalized signal intensity in the K562 RNA sample, negative M values indicate higher intensity in the pool RNA sample, and M values of zero indicate equal intensity in the two samples. A is a log2-transformed measure of total signal intensity for both samples. Higher A values indicate brighter signals. M and_A_ values for all long oligonucleotide probes are shown inFigure 1A, B. These M and A values are means of six replicate arrays. Compared with the human gene probes, randomized negative control long oligonucleotide probes all produced dim signals (low A values) and M values close to zero. For example, for the Version 1 arrays, the 29 randomized negative control probes had A values of 6.8 ± 0.2 and M values of 0.08 ± 0.12 (mean ± standard deviation).
Figure 1.
Differential expression and signal intensity measurements for all three array types. (A) Version 1 spotted long oligonucleotide arrays (means from six replicate two-color hybridizations). (B) Version 2 spotted long oligonucleotide arrays (four replicate two-color hybridizations). (C) In situ-synthesized 25-mer arrays (two replicate K562 sample single-color hybridizations and three replicate pool sample single-color hybridizations). Each point represents data from a single long oligonucleotide probe (A, B) or 25-mer probe set (C). M is a measure of differential gene expression (log2 [K562 intensity / pool intensity]). A is a measure of signal intensity (0.5 log2 K562 intensity + 0.5 log2 pool intensity).
We analyzed the same two RNA samples using in situ-synthesized 25-mer oligonucleotide arrays. We analyzed K562 and pool RNA samples separately, because this technology has been optimized for single-color hybridizations. Normalized log2-transformed absolute signal intensities were calculated for each probe set on each array using RMA software. After averaging across replicate hybridizations, intensity values from K562 and pool arrays were combined to calculate M and A values for each probe set (Fig. 1C). Absolute signal intensities (A values) differ between long oligonucleotide arrays and 25-mer arrays, because these values were calculated from raw data obtained using different technologies. When we used another algorithm (Affymetrix MAS 5) that compares signal intensity from perfect-match and mismatch 25-mers, about half of the genes surveyed on these arrays were called “present” in these samples. For example, for the three replicate arrays used to analyze the pool sample,48%–53% of probe sets were called “present,” 45%–49% “absent,” and the remainder “marginal.”
We compared measurements obtained using the three different array types. Because probe designs were based on different sets of GenBank cDNA sequences, we assigned each long oligonucleotide probe or 25-mer probe set to a cluster using the UniGene database (Build 155). The numbers of Uni-Gene clusters represented in each probe group are listed inTable 1. The distribution of UniGene clusters between the three array types is shown inFigure 2. There were 7344 gene clusters that were represented on each of the three array types we used. We used this large set of genes as a basis for comparing expression data from the three array types.
Figure 2.
Representation of genes on the three array types. Figures indicate the numbers of genes (distinct UniGene clusters) represented by at least one probe (or probe set) on each of the array types employed in this study. A total of 7344 genes were represented on all three array types.
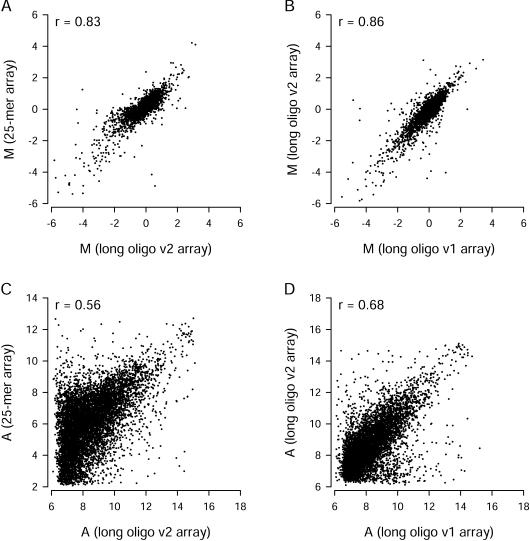
Expression data from Version 1 spotted long oligonucleotide arrays and in situ-synthesized 25-mer arrays are compared inFigure 3. When all 7344 clusters were included in the comparison, there was a clear correlation between M values obtained using the two array types (Fig. 3A, Pearson correlation coefficient r = 0.80). The magnitudes of M values were similar for the two array types. We considered the effect of signal intensity on the _M_-value comparison. Some long oligonucleotide probes gave signals that were in the same range seen for negative control probes, and the 25-mer probe sets with low signals were usually called “absent” by the MAS 5 algorithm. When we excluded measurements from all probes and probe sets associated with A values less than the median A value for all probes on the array, the correlation between M values for the 2877 remaining genes improved substantially (r = 0.89,Fig. 3B). A very similar correlation was obtained when we reanalyzed the arrays with Affymetrix MAS 5 (for 25-mer arrays) and Axon GenePix 3 (for long oligonucleotide arrays), and then excluded 25-mer data associated with “absent” calls and long oligonucleotide data associated with “not found” spots (data not shown). However, that approach was more difficult to implement because it required manual spot flagging, and special rules were needed to handle missing values from individual arrays. The A values from the two array types were not as highly correlated as the M values (Fig. 3C). To help compare variance within the Version 1 long oligonucleotide array replicates with between-platform variance, we correlated average M values from the first three replicates with average M values from the last three replicates (Fig. 3D). As expected, the within-platform correlation (r = 0.94) was somewhat higher than the correlations obtained for the cross-platform comparison (Fig. 3A, B). We found similar cross-platform correlations when we compared the Version 2 long oligonucleotide arrays to the other two array types (Fig. 4). For all comparisons, there was a clear correlation between differential expression measurements made with different array types, and the correlation improved when measurements from probes with low intensity signals were excluded.
Figure 3.
Comparison of gene expression measurements from Version 1 (v1) long oligonucleotide arrays and in situ-synthesized 25-mer arrays. (A) Differential gene expression measurements for all 7344 common genes. (B) Differential gene expression measurements for 2877 genes remaining after exclusion of probes or probes sets with low signal intensities (A values below the median for that array type). (C) Comparison of signal intensities for all 7344 common genes. (D) Comparison of M values obtained from the first three and the last three v1 oligonucleotide arrays. Each point represents data from a single UniGene cluster. Pearson correlation coefficients (r) are shown for each comparison. Dashed lines are lines of equality.
Figure 4.
Comparison of gene expression measurements on Version 2 (v2) long oligonucleotide arrays. Differential expression (A, B) and signal intensity (C, D) measurements from these arrays were compared with measurements from in situ-synthesized 25-mer arrays (A, C) and Version 1 long oligonucleotide arrays (B, D). Comparisons involve the 7344 common genes. For M value comparisons, probes or probe sets with low signal intensity were excluded. After exclusion of those probes, a total of 3133 genes (A) or 3344 genes (B) remained for comparison.
Investigators are often interested in identifying highly differentially expressed genes. To examine how genes with high differential expression measurements compare on the three array types tested, we generated a list of all genes that produced one of the 10 highest or 10 lowest M values on any of the three array types (Table 2). All 7344 gene clusters represented on each of the three array types were considered. By this criterion, a total of 22 genes were found to have extremely high relative expression in K562 RNA (M value in the top 10 on at least one array type). Of these 22 genes,16 were among the top 2% of M values on all three array types, four more genes were among the among the top 2% on 25-mer arrays and one of the spotted long oligonucleotide arrays, and two genes (VCY and COLEC10) were only found to be substantially differentially expressed on the Version 2 long oligonucleotide arrays. The findings were very similar for the 22 total genes found to have one of the 10 lowest M values on at least one array type: 16 were in the bottom 2% on all three array types, five were in the bottom 2% on 25-mer arrays and one of the spotted long oligonucleotide arrays, and one (MT2A) appeared to be highly differentially expressed only on the Version 1 long oligonucleotide array. Of all 44 genes listed in Table 2, there were 12 genes that did not give measurements in the top (or bottom) 2% on at least one array type (rank >2% or <98%). In 11 of these 12 cases, the failure to detect substantial differential expression was associated with low signal intensity or probes predicted to recognize different splice variants (see Table 2).
Table 2.
Highly Differentially Expressed Genes
| 25-mer arrays | Long oligo v1 arrays | Long oligo v2 arrays | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UniGene cluster | Symbol | Gene description | M | Fold diff.a | Rankb | M | fold diff.a | Rankb | M | Fold diff.a | Rankb |
| Among 10 most highly differentially expressed in K562 RNA sample on at least one array type | |||||||||||
| Hs. 2841 | NMU | Neuromedin U | 4.10 | 17.2 | <1% | 3.43 | 10.8 | <1% | 3.14 | 8.8 | <1% |
| Hs. 289105 | SSX2 | Synovial sarcoma, X breakpoint 2 | 4.47 | 22.2 | <1% | 2.58 | 6.0 | <1% | 2.91 | 7.5 | <1% |
| Hs. 128231 | GAGEB1 | G antigen, family B, 1 | 2.99 | 7.9 | <1% | 2.99 | 7.9 | <1% | 2.58 | 6.0 | <1% |
| Hs. 169536 | RHAG | Rhesus blood group-assoc glycoprotein | 2.56 | 5.9 | <1% | 2.18 | 4.5 | <1% | 2.32 | 5.0 | <1% |
| Hs. 37860 | KLF1 | Kruppel-like factor 1 (erythroid) | 2.93 | 7.6 | <1% | 2.32 | 5.0 | <1% | 1.72 | 3.3 | <1% |
| Hs. 1861 | MPP1 | Membrane protein, palmitoylated 1, 55 kDa | 2.46 | 5.5 | <1% | 1.87 | 3.7 | <1% | 2.40 | 5.3 | <1% |
| Hs. 99962 | PRG2 | Proteoglycan 2, bone marrow | 2.01 | 4.0 | <1% | 1.99 | 4.0 | <1% | 2.69 | 6.5 | <1% |
| Hs. 77439 | PRKAR2B | cAMP-dependent protein kinase reg 2 beta | 2.44 | 5.4 | <1% | 2.04 | 4.1 | <1% | 2.15 | 4.4 | <1% |
| Hs. 184585 | LMO2 | LIM domain only 2 (rhombotin-like 1) | 2.81 | 7.0 | <1% | 1.59 | 3.0 | <1% | 2.22 | 4.7 | <1% |
| Hs. 188021 | KCNH2 | Potassium voltage-gated channel H2 | 2.05 | 4.2 | <1% | 1.71 | 3.3 | <1% | 2.73 | 6.6 | <1% |
| Hs. 372513 | GYPB | Glycophorin B (includes Ss blood group) | 2.33 | 5.0 | <1% | 1.84 | 3.6 | <1% | 2.32 | 5.0 | <1% |
| Hs. 30743 | PRAME | Preferentially expressed antigen in melanoma | 2.23 | 4.7 | <1% | 2.13 | 4.4 | <1% | 2.10 | 4.3 | <1% |
| Hs. 78601 | UROD | Uroporphyrinogen decarboxylase | 2.21 | 4.6 | <1% | 1.99 | 4.0 | <1% | 2.02 | 4.0 | <1% |
| Hs. 1985 | SPTA1 | Spectrin, alpha, erythrocytic 1 (elliptocytosis 2) | 2.89 | 7.4 | <1% | 1.00 | 2.0 | <2% | 2.17 | 4.5 | <1% |
| Hs. 3132 | STAR | Steroidogenic acute regulatory protein | 2.95 | 7.7 | <1% | 1.47 | 2.8 | <1% | 1.59 | 3.0 | <1% |
| Hs. 158213 | SPAG6 | Sperm associated antigen 6 | 2.93 | 7.6 | <1% | 1.19 | 2.3 | <1% | 1.52 | 2.9 | <1% |
| Hs. 194759 | SSX1 | Synovial sarcoma, X breakpoint 1 | 2.39 | 5.3 | <1% | -0.37 | -1.3 | >2%c | 3.11 | 8.6 | <1% |
| Hs. 765 | GATA1 | GATA binding protein 1 (globin transcript. factor 1) | 2.36 | 5.1 | <1% | 2.43 | 5.4 | <1% | 0.00 | -1.0 | >2%c |
| Hs. 178749 | SSX3 | Synovial sarcoma, X breakpoint 3 | 2.78 | 6.8 | <1% | 1.17 | 2.2 | <1% | 0.04 | 1.0 | >2%d |
| Hs. 176660 | GAGE1 | G antigen 1 | 2.79 | 6.9 | <1% | 1.11 | 2.2 | <1% | 0.03 | 1.0 | >2%d |
| Hs. 170076 | VCY | Variable charge, Y chromosome | 0.06 | 1.0 | >2%c | 0.08 | 1.1 | >2%c | 2.51 | 5.7 | <1% |
| Hs. 176615 | COLEC10 | Collectin subfamily member 10 (C-type lectin) | 0.02 | 1.0 | >2%d | -0.20 | -1.1 | >2%d | 2.50 | 5.7 | <1% |
| Among 10 most highly differentially expressed in pool RNA sample on at least one array type | |||||||||||
| Hs. 287820 | FN1 | Fibronectin 1 | -5.78 | -54.8 | >99% | -5.46 | -44.0 | >99% | -6.44 | -86.6 | >99% |
| Hs. 350470 | TFF1 | Trefoil factor 1 | -5.29 | -39.1 | >99% | -5.54 | -46.5 | >99% | -5.58 | -47.8 | >99% |
| Hs. 93194 | APOA1 | Apolipoprotein A-I | -4.36 | -20.6 | >99% | -4.28 | -19.4 | >99% | -5.74 | -53.3 | >99% |
| Hs. 111779 | SPARC | Secreted protein, acidic, cysteine-rich | -4.22 | -18.6 | >99% | -4.53 | -23.2 | >99% | -5.12 | -34.8 | >99% |
| Hs. 1252 | APOH | Apolipoprotein H (beta-2-glycoprotein I) | -4.69 | -25.9 | >99% | -3.54 | -11.6 | >99% | -5.43 | -43.1 | >99% |
| Hs. 324746 | AHSG | Alpha-2-HS-glycoprotein | -4.77 | -27.2 | >99% | -3.99 | -15.9 | >99% | -4.90 | -29.8 | >99% |
| Hs. 181244 | HLA-A | Major histocompatibility complex, class I, A | -5.40 | -42.1 | >99% | -3.59 | -12.0 | >99% | -4.67 | -25.5 | >99% |
| Hs. 74034 | CAV1 | Caveolin 1 | -5.35 | -40.8 | >99% | -4.01 | -16.1 | >99% | -4.00 | -16.0 | >99% |
| Hs. 137476 | PEG10 | Paternally expressed 10 | -5.50 | -45.3 | >99% | -3.76 | -13.5 | >99% | -4.07 | -16.8 | >99% |
| Hs. 284176 | TF | Transferrin | -4.69 | -25.9 | >99% | -3.38 | -10.4 | >99% | -4.86 | -29.1 | >99% |
| Hs. 289114 | TNC | Tenascin C (hexabrachion) | -4.82 | -28.3 | >99% | -3.05 | -8.3 | >99% | -4.87 | -29.3 | >99% |
| Hs. 95972 | SILV | Silver homolog (mouse) | -3.10 | -8.5 | >99% | -5.14 | -35.4 | >99% | -4.45 | -21.8 | >99% |
| Hs. 119651 | GPC3 | Glypican 3 | -4.87 | -29.3 | >99% | -3.17 | -9.0 | >99% | -3.98 | -15.8 | >99% |
| Hs. 118787 | TGFBI | Transforming growth factor, beta-induced, 68 kDa | -3.94 | -15.4 | >99% | -2.89 | -7.4 | >99% | -4.63 | -24.7 | >99% |
| Hs. 1741 | ITGB7 | Integrin, beta 7 | -2.26 | -4.8 | >98% | -4.04 | -16.5 | >99% | -4.46 | -22.1 | >99% |
| Hs. 75431 | FGG | Fibrinogen, gamma polypeptide | -4.96 | -31.1 | >99% | -2.63 | -6.2 | >99% | -2.05 | -4.1 | >98% |
| Hs. 585 | APOB | Apolipoprotein B | -4.88 | -29.5 | >99% | -4.83 | -28.4 | >99% | 0.57 | 1.5 | <98%c |
| Hs. 621 | LGALS3 | Lectin, galactoside-binding, soluble, 3 (galectin 3) | -4.54 | -23.2 | >99% | -4.32 | -20.0 | >99% | 0.25 | 1.2 | <98%d |
| Hs. 750 | FBN1 | Fibrillin 1 (Marfan syndrome) | -3.52 | -11.5 | >99% | -4.39 | -21.0 | >99% | 0.03 | 1.0 | <98% |
| Hs. 1244 | CD9 | CD9 antigen (p24) | -4.94 | -30.7 | >99% | 0.01 | 1.0 | <98%d | -2.69 | -6.5 | >98% |
| Hs. 41270 | PLOD2 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | -4.97 | -31.3 | >99% | -0.01 | -1.0 | <98%d | -2.36 | -5.2 | >98% |
| Hs. 118786 | MT2A | Metallothionein 2A | -0.57 | -1.5 | <98%c | -4.41 | -21.3 | >99% | -0.71 | -1.6 | <98%c |
Graphical comparisons involving larger groups of genes with extreme_M_ values are shown in Figure 5. Genes found to have M values near zero (equal expression in the two RNA samples) using one array type were highly unlikely to have large M values on another array (Fig. 5B). For example, of the 5676 genes with 25-mer array M values between -0.5 and 0.5, the Version 1 long oligonucleotide array results showed that 89% had M values between -0.5 and 0.5 and 99% had M values between -1 and 1 (i.e., less than twofold difference). Most genes associated with the highest and lowest M values on 25-mer arrays gave similar values on long oligonucleotide arrays (Fig. 5B, C). However, long oligonucleotide arrays did tend to give somewhat smaller estimates for the magnitude of M for genes with the most negative M values (Fig. 5C). In summary, genes producing extreme M values on one array type usually also produced large M values on the other two array types, whereas genes that produced small M values on one array type rarely produced large M values on another.
Figure 5.
Differential expression measurements for genes with the highest and lowest_M_ values on 25-mer arrays. Genes were divided into subsets according to the M values determined on 25-mer arrays. (A) Genes with_M_ values in the top 1%. (B) Genes with M values close to zero. (C) Genes with M values in the bottom 1%. Each panel shows M values for these subsets of genes as measured using 25-mer arrays (solid lines), Version 1 long oligonucleotide arrays (dotted lines), and Version 2 long oligonucleotide arrays (dashed lines).
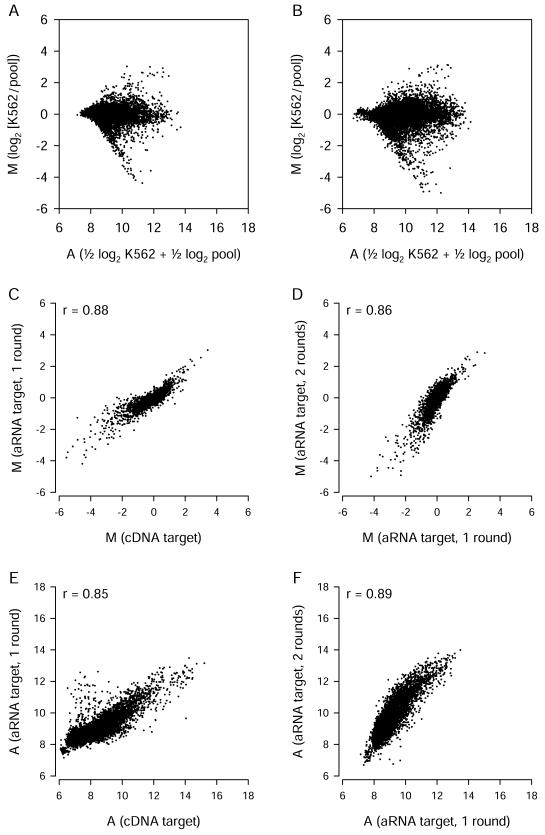
We also examined whether spotted oligonucleotide arrays could be used successfully with amplified targets. In preliminary experiments involving unamplified cDNA targets, we found that ∼20 μg of total RNA was required for optimal signal intensity. To allow for the use of spotted arrays with smaller RNA samples, we used a modification of the Eberwine T7 RNA polymerase-based method (Eberwine et al. 1992) to produce cRNA. Starting with 1 μg of total RNA for one-round amplifications or 5–10 ng of total RNA for two-round amplifications, we obtained sufficient cRNA for multiple hybridizations. Amplified targets generally produced somewhat less intense fluorescence. (Because we used higher photomultiplier tube voltages to scan the arrays, this did not result in lower A values.) Mean M and A values obtained using cRNA produced by one or two rounds of amplification are shown in Figure 6A, B.
Figure 6.
Amplified targets used with Version 1 spotted long oligonucleotide arrays. (A) Expression data from six replicate arrays hybridized with cRNA targets produced using a single round of amplification. (B) Data from two replicate arrays with cRNA targets produced using two rounds of amplification. (C, D) Comparisons of M values obtained using unamplified and amplified targets, after exclusion of probes with low_A_ values. After exclusion of low-intensity signals, 5746 probes (C) or 5771 probes (D) of 13,971 total probes remained for comparison. (E, F) Signal intensity values for unamplified and amplified targets. Each point represents data from a single probe.
We compared estimates of differential gene expression (M) made using cDNA targets and cRNA targets. For consistency, we limited this analysis to the same set of 7344 gene clusters and used the same strategy to filter out low-intensity signals. The cDNA target M values correlated well with one-round cRNA target M values, but the cRNA targets tended to result in smaller estimates of the extent of differential gene expression (Fig. 6C). M values were maintained within a similar range following a second round of amplification (Fig. 6D). Comparisons of A values from unamplified and amplified target hybridizations indicate that a small subset of probes produced higher_A_ values than expected when amplified targets were used (Fig. 6E). These probes may bind nonspecifically to cRNA targets or recognize transcripts that are unusually highly amplified. In contrast, there were very few probes that produced A values dramatically lower than expected. Although the amplification procedure is expected to bias transcript representation to a certain extent, these results suggest that most transcripts that can be measured using unamplified cDNA targets can also be measured using amplified cRNA targets. When used with spotted long oligonucleotide arrays, cRNA targets provided estimates of differential gene expression (M) that correlated well with those obtained using cDNA targets, although the magnitude of M was typically smaller.
DISCUSSION
Most reported evaluations of DNA microarray platforms have focused on analyzing expression measurements made on a relatively small fraction of the many thousands of genes represented on modern arrays. In those studies, microarray measurements were calibrated using known amounts of input RNAs or were compared to measurements made with another technique, such as quantitative PCR. These approaches have yielded important information about the sensitivity, specificity, and reproducibility of these platforms, at least for the subset of genes studied directly. We took a different approach that allowed us to compare thousands of gene expression measurements made using a total of five different combinations of array types and target preparation methods. By using each method to compare gene expression in the same pair of dissimilar RNA samples, our approach provided a wide range of expression ratios for comparison and made it possible to examine how various sets of measurements made with spotted 70-mer oligonucleotide arrays correlated with one another and with measurements made using in situ-synthesized 25-mer arrays. Our ability to compare results between different data sets, including those based on single-color as well as two-color hybridizations, was facilitated by expressing each measurement as a pair of log-transformed differential expression (M) and total signal (A) values. The general approach described here should prove useful for the ongoing evaluation of new methods for genome-scale gene expression analysis.
Given the serious concerns raised in some previous comparisons of different microarray platforms (Kothapalli et al. 2002; Kuo et al. 2002; Li et al. 2002), we were pleased to find very good overall agreement among the methods we examined here. We found that two groups of differential expression measurements made with separate collections of long oligonucleotide probes were clearly correlated with each other and with a third group of measurements made using in situ-synthesized 25-mer probe sets. Because expression measurements associated with low signal intensities are known to be less reliable (Yang et al. 2001), it is not surprising that the strength of the correlation between differential expression measurements improved when genes with low-intensity signals on either array type were excluded from the comparison. We also found good correlations between long oligonucleotide array measurements made with unamplified (cDNA) targets and with amplified (cRNA) targets. In other experiments involving murine RNA samples, we found that spotted long oligonucleotide array data agreed well with quantitative real-time PCR data for a small subset (40 genes) of the genes represented on those arrays (D.J. Erle, unpubl.). Our results suggest that spotted long oligonucleotide arrays should be useful for large-scale gene expression analysis in humans and other complex organisms.
Spotted arrays are generally used for two-color hybridizations, and many study designs involve comparison of each test sample to a common reference sample. This design has been successfully employed for many spotted cDNA array experiments. To allow for accurate quantification of a particular gene, the reference sample must contain sufficient RNA to produce a clear signal for the corresponding probe. Reference samples are often generated from a pool of different cell lines, as was one of the two RNA samples analyzed here. Using Affymetrix MAS 5 software to analyze our three replicate 25-mer array analyses of the pool sample, we found that only 48%–53% of the probe sets yielded “present” calls (with the remainder being judged as either “marginal” or “absent”). More importantly, we found that a substantial fraction of 70-mer probes were associated with signal intensities that were no brighter than those seen for randomized negative control probes. These results suggest that reference pools such as the one used here may not produce sufficient signal to allow for accurate quantification of some genes on spotted long oligonucleotide arrays. If this is an important issue, the use of different reference samples with gene expression levels similar to those found in the test samples or the use of reference-free designs (where pairs of test samples are compared directly) may be preferable.
Our results indicate that probe selection can have important effects on net signal intensity and on measurements of differential gene expression. When we compared two different collections of ∼70-mer probes, we sometimes found dramatic signal intensity (A value) differences between two probes that corresponded to the same gene (Fig. 4D). M values were generally more closely related, although there were exceptions (Fig. 4B,Table 2). Variability in A and M values was also seen when different 25-mer probe sets associated with the same GenBank accession number or UniGene cluster were compared (data not shown). These between-probe differences may be attributable to a number of factors, including alternative splicing, probe GC content, nucleic acid structure, and distance from the 3′ end of the RNA transcript, as well as by the amount of probe available for hybridization. Because our comparisons depend upon UniGene, it is also possible that between-probe differences could result from inappropriate assignment of GenBank sequences from different genes to the same UniGene cluster. Despite all of these potential concerns, we found good agreement between in situ-synthesized oligonucleotide arrays with sets of multiple short oligonucleotides for every gene and spotted arrays containing just a single long oligonucleotide for most genes. We simulated the performance of arrays with two long oligonucleotides per gene by averaging together the M values obtained with the two different oligonucleotide collections, Versions 1 and 2. This led to only a trivial improvement in the overall correlation between long oligonucleotide array M values and 25-mer array_M_ values (data not shown). On the other hand, we found some cases in which two long oligonucleotides designed to recognize the same gene gave discordant results, probably due at least in part to alternative splicing and to low sensitivity of some probes (Table 2,Fig. 4). Whether the potential benefits of additional probes justify the additional cost and complexity will depend upon the intended application. In any case, because probe selection has important effects, the adoption of standardized sets of probes with publicly available sequences will be important for investigators wishing to generate data that can be compared with data from other studies (Li and Stormo 2001;Wright and Church 2002).
Selection of a suitable microarray platform for a specific application can be influenced by a number of considerations. Many investigators have used in situ-synthesized 25-mer arrays, but the application of this technology has been limited by cost considerations, especially for projects involving large numbers of samples. Spotted arrays can be produced in quantity by individual laboratories or core facilities at a lower cost, although this can be labor-intensive and considerable expertise is required. Most spotted arrays have been made with probes made from cDNA clones, and much of the effort required to produce spotted cDNA arrays has centered on obtaining, sequence-verifying, and amplifying suitable cDNA libraries. In contrast, collections of long oligonucleotides are available from various sources, and sequence verification and amplification are not required. The overall costs of long oligonucleotide technology will often be lower when labor and other costs associated with obtaining and maintaining cDNA libraries are taken into account, and spotted long oligonucleotide arrays can be printed, hybridized, and scanned using virtually the same methods and equipment used for spotted cDNA arrays. Furthermore, long oligonucleotide probes can be designed to have more uniform hybridization characteristics and to avoid sequences with a high degree of homology to other genes; and probes for novel genes, gene variants, and transgenes can be designed using freely available tools and added easily to existing probe collections. Our large-scale analysis showed a strong correlation between spotted long oligonucleotide array data and in situ-synthesized 25-mer array data, suggesting that long oligonucleotide arrays are a good alternative gene expression analysis platform for many applications.
METHODS
Spotted Array Fabrication
Long oligonucleotides were designed and synthesized by Operon. Operon provided the following information about the oligonucleotides: Human Genome Oligo Set Version 1 included 13,971 oligonucleotides, mostly 70-mers, designed based upon representative sequences in build 119 of the human UniGene database. Human Genome Oligo Set Version 2 included 21,329 oligonucleotides, mostly 69-mers, that were designed based upon UniGene build 147. An amino linker was attached to the 5′ end of each oligonucleotide. Oligonucleotides were designed to have melting temperatures of 78°C ± 5°C using the formula Tm = 81.5 + 16.6 × log [Na+] + 41 × %GC - 500/length, where [Na+] = 0.1 M. The GC content (%GC) was 48% ± 6% (mean±sd) for Version 1 and 49% ± 5% for Version 2. For Version 1,>99% of the probes were 70-mers, whereas for Version 2,>99% were 69-mers. In a few cases, probe length was adjusted to keep Tm within the desired range. Probes were 3′ biased: ∼96% of the Version 1 probes and 99% of the Version 2 probes were within 600 bases of the 3′ end of the known sequence, although some sequences were incomplete (lacked a polyA sequence). BLAST searches were done to exclude probes that cross-hybridized with other sequences from the UniGene database. Ninety percent of the Version 1 probes had less than 85% overall identity with any other sequence, and 95% of the Version 2 probes had less than 54% overall identity with any other sequence. Both sets also included randomized negative oligonucleotides.
Oligonucleotides were dissolved in 3× SSC at a concentration of 40 μM in preparation for spotting on poly-L-lysine-coated glass slides, prepared as described athttp://www.microarrays.org. A custom-built microarrayer equipped with Majer Precision Engineering MicroQuill pins was used to print each batch of 255 slides. A detailed description of the microarrayer is available athttp://cmgm.stanford.edu/pbrown. After printing, the slides were stored at room temperature in a desiccator.
RNA Samples
K562 erythroleukemia cells (American Type Culture Collection, CCL-243) were cultured in RPMI 1640 medium with L-glutamine (Mediatech) supplemented with 10% fetal bovine serum (HyClone),100 IU/mL penicillin (Mediatech), and 100 μg/mL streptomycin (Mediatech). Cells were maintained at 37°C under 5% CO2/95% air in a humidified incubator. Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. A pooled RNA sample derived from 10 human cell lines was purchased from Stratagene (Universal Human Reference RNA).
Preparation of Labeled cDNA Targets for Spotted Oligonucleotide Arrays
Reverse transcription reactions were performed as described by DeRisi et al. (1997) with modifications. Briefly, total RNA (20 μg) and oligo-dT (2 μg) were incubated at 70°C for 10 min and snap-cooled on ice. Superscript II reverse transcriptase (600 U) and Superscript II reaction buffer (Invitrogen); dATP, dCTP, and dGTP (0.5 mM each),0.2 mM dUTP, and 0.3 mM aminoallyl-dUTP (Sigma-Aldrich) were added, and the mixture was incubated at 42°C for 2 h. RNA was degraded by the addition of NaOH (0.2 N final concentration) and EDTA (0.1 M final concentration), followed by incubation at 65°C for 15 min. The reaction was neutralized by the addition of HEPES pH 7.0. The cDNAs were desalted and concentrated using a Microcon-30 concentration unit (Millipore) and then lyophilized. After resolubilization in 0.1 M sodium bicarbonate pH 9.0, cDNAs were coupled to N-hydroxysuccinimidyl esters of Cy3 or Cy5 dyes (CyScribe, Amersham Biosciences) for 1 h in the dark. Coupling reactions were quenched with 9 μL of 4M hydroxylamine. After addition of 35 μL of 0.1M sodium acetate pH 5.2, Cy3- and Cy5-labeled cDNAs were combined and co-purified using a Qiaquick column (QIAGEN).
Spotted Oligonucleotide Array Hybridization and Scanning
Prior to hybridization, excess oligonucleotide was removed from the arrays by gently shaking them for 10 min in 0.2% SDS. Arrays were then washed 5 times with distilled, filtered water and dried for 5 min at 600 rpm in a centrifuge. A solution of 0.1% SDS,5× SSC and 1% BSA was applied to the arrays for 30 min at 42°C to block nonspecific binding. Arrays were washed 5 times with distilled, filtered water and dried for 5 min at 600 rpm in a centrifuge. Labeled cDNAs were added to hybridization buffer (2.8× SSC,0.025 M HEPES pH7.0, 0.2% SDS,1 μg/μL yeast tRNA, and 0.6 μg/μL human Cot-1 DNA), denatured for 5 min at 95°C, and applied to the arrays using a cover slip (LifterSlip, Erie Scientific). Arrays were placed in a Dietech hybridization chamber, and chambers were immersed in a 63°C water bath for 20 h. After hybridization, arrays were washed successively with 1× SSC with 0.03% SDS,0.2× SSC, and 0.05× SSC for 3 min each at room temperature and then dried. Arrays were scanned using an Axon 4000B laser scanner.
Spotted Oligonucleotide Array Image Analysis
For spotted oligonucleotide arrays, image analyses were performed using the software package Spot (Yang et al. 2002a). For each element, Cy5 (red) and Cy3 (green) signal intensities were corrected by subtracting the morphological opening background intensity from the foreground fluorescence intensity. Corrected signal intensities were used to calculate M and A values for every element on each array. M, a measure of differential gene expression, was calculated as log2 (K562 intensity/pool intensity). A, a measure of signal strength, was calculated as 0.5 log2 K562 intensity + 0.5 log2 pool intensity. M values were normalized using the R package SMA by the within print tip group lowess normalization procedure (Ihaka and Gentleman 1996; Yang et al. 2002b).
Measurement of Gene Expression Using In Situ-Synthesized 25-mer Arrays
Double-stranded cDNA was synthesized from 6–11 μg of total RNA with the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) and dT24-T7 primer (Operon) according to the manufacturers' instructions. Biotin-labeled cRNA was prepared by in vitro transcription using the BioArray High Yield RNA Transcript Labeling Kit (Enzo). Fragmentation of cRNA, hybridization, washing, staining, and scanning were performed as described in the Affymetrix GeneChip Expression Analysis Technical Manual. In brief, biotinylated cRNA was fragmented to 35–200 base pairs with fragmentation buffer (40 mM Tris-acetate pH 8.1,100 mM potassium acetate,30 mM magnesium acetate) for 35 min at 94°C. For each sample, 30 μg of fragmented, biotinylated cRNA was combined with prelabeled eukaryotic hybridization controls (Affymetrix). The cRNA samples were first hybridized to Affymetrix Test3 arrays for quality control and subsequently to Affymetrix HG-U95Av2 arrays. These arrays include ∼16 probe pairs (perfect-match and mismatch 25-mers) per probe set. To estimate the GC content of these 25-mer probes, we randomly selected eight probe sets containing a total of 128 probes. The GC content was 44% ± 8% (mean±sd). Most probes in these sets were within ∼600 bases of the 3′ end of the target sequence. Hybridizations were carried out at 45°C for 16 h in a rotisserie at 60 rpm. Following hybridization, the arrays were washed and stained in the Affymetrix Fluidics Station 400, using an antibody amplification protocol and streptavidin-phycoerythrin. Arrays were scanned with the Affymetrix GeneArray Scanner. Expression values were calculated using the Robust Multichip Average (RMA) method (Irizarry et al. 2003b, Irizarry et al.,2003a). RMA estimates are based upon a robust average of background corrected PM intensities. Normalization was done using quantile normalization (Bolstad et al. 2003). The model was fitted using the iteratively reweighted least squares (IRLS) algorithm with Tukey's bi-square using the function rlm() from the R software package (available athttp://www.r-project.org). Log2-transformed intensity values were averaged across replicate arrays and used to calculate M and A values. M was calculated as log2 (K562 intensity) - log2 (pool intensity). A was calculated as 0.5 log2 (K562 intensity) + 0.5 log2 (pool intensity).
Target Amplification for Spotted Oligonucleotide Arrays
RNA amplifications were performed using the MessageAmp aRNA Kit (Ambion) according to the manufacturer's protocol. One μg of total RNA was used for one-round amplification experiments, and 5–10 ng of total RNA was used for two-round amplifications. Amino-allyl UTP (Sigma-Aldrich) was incorporated into the final cRNA product. Yields of cRNA were 15–40 μg (one round) and 40–45 μg (two rounds). We used 3–7.5 μg of cRNA per sample for each array. Coupling to Cy3 and Cy5 was performed with the same method used for cDNA coupling (see above). Labeled cRNA targets were purified using the RNeasy Mini Prep Kit (QIAGEN) and fragmented with the same method used for biotinylated cRNAs (see above). Hybridization, washing, scanning, and image analysis were performed using the same methods described for cDNA targets.
Cross-Platform Comparisons of Expression Data
To facilitate comparisons between data sets, long oligonucleotide probes and 25-mer probe sets were mapped to clusters according to the human UniGene database, build 155. GenBank accession numbers provided by the manufacturers (Operon and Affymetrix) were submitted to the Stanford Online Universal Resource for Clones and ESTs (SOURCE,http://source.stanford.edu) to obtain UniGene cluster assignments. In some cases, a single UniGene cluster was represented by more than one probe (or probe set) on a single array. This occurred most commonly for the in situ-synthesized 25-mer arrays, and was relatively uncommon for the long oligonucleotide arrays (seeTable 1). When there was more than one probe for a cluster, M and A values for that cluster were determined by averaging the corresponding probes or probe sets. For some comparisons, expression data were filtered to remove low-intensity signals (A values below the median for all probes on the array). The relationships between expression measurements made with different array types were assessed by Pearson correlation coefficients.
Acknowledgments
We thank Tanuja Goulet and Michael Salazar for technical assistance and Chris Barker and Dean Sheppard for helpful discussions. This work was funded by the UCSF Sandler Center for Basic Research in Asthma, by NIH grants HL072301, DK54212, and RR00083, and by an NHLBI-funded BayGenomics Program in Genomic Applications (HL66621, HL66600, and HL66590).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1048803.
Footnotes
Article published online before print in June 2003.
[Data from this study are available from GEO (http://www.ncbi.nlm.nih.gov/geo)and are listed under the following accession numbers: GSE344 (for the entire experimental series), GSM4843-GSM4865 (for the expression data from individual arrays), and GPL91, GPL273, and GPL274 (for the three array platforms).]
References
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19**:** 185-193. [DOI] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278**:** 680-686. [DOI] [PubMed] [Google Scholar]
- Eberwine, J., Yeh, H., Miyashiro, K., Cao, Y., Nair, S., Finnell, R., Zettel, M., and Coleman, P. 1992. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. 89**:** 3010-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.R., Mao, M., Jones, A.R., Burchard, J., Marton, M.J., Shannon, K.W., Lefkowitz, S.M., Ziman, M., Schelter, J.M., Meyer, M.R., et al. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19**:** 342-347. [DOI] [PubMed] [Google Scholar]
- Ihaka, R. and Gentleman, R. 1996. R: A language for data analysis and graphics. J. Comp. Graph. Stat. 5**:** 299-314. [Google Scholar]
- Irizarry, R.A., Hobbs, B., Colin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. 2003a. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics (in press). [DOI] [PubMed]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. 2003b. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31**:** e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, M.D., Jatkoe, T.A., Stumpf, C.R., Lu, J., Thomas, J.D., and Madore, S.J. 2000. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 28**:** 4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli, R., Yoder, S.J., Mane, S., and Loughran Jr., T.P. 2002. Microarray results: How accurate are they? BMC Bioinformatics 3**:** 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, W.P., Jenssen, T.K., Butte, A.J., Ohno-Machado, L., and Kohane, I.S. 2002. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics 18**:** 405-412. [DOI] [PubMed] [Google Scholar]
- Li, F. and Stormo, G.D. 2001. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics 17**:** 1067-1076. [DOI] [PubMed] [Google Scholar]
- Li, J., Pankratz, M., and Johnson, J.A. 2002. Differential gene expression patterns revealed by oligonucleotide versus long cDNA arrays. Toxicol. Sci. 69**:** 383-390. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J., Dong, H., Byrne, M.C., Follettie, M.T., Gallo, M.V., Chee, M.S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., et al. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14**:** 1675-1680. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. 1996. Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. 93**:** 10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, M.A. and Church, G.M. 2002. An open-source oligomicroarray standard for human and mouse. Nat. Biotechnol. 20**:** 1082-1083. [DOI] [PubMed] [Google Scholar]
- Yang, M.C., Ruan, Q.G., Yang, J.J., Eckenrode, S., Wu, S., McIndoe, R.A., and She, J.X. 2001. A statistical method for flagging weak spots improves normalization and ratio estimates in microarrays. Physiol. Genomics. 7**:** 45-53. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Buckley, M.J., Dudoit, S., and Speed, T.P. 2002a. Comparison of methods for image analysis on cDNA microarray data. J. Comp. Graph. Stat. 11**:** 108-136. [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. 2002b. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30**:** e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen, T., Wurmbach, E., Pfeffer, R.L., Ebersole, B.J., and Sealfon, S.C. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30**:** e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/geo; Gene Expression Omnibus, NCBI, a public repository for expression data, including the data reported here.
- http://www.microarrays.org; DeRisi laboratory, University of California, San Francisco, a public source for microarray protocols and software.
- http://www.r-project.org; The R Project for Statistical Computing, describes and provides access to R, used for data analyses presented here.
- http://source.stanford.edu; Stanford Online Universal Resource for Clones and ESTs, provided a tool for batch assignment of UniGene cluster identifiers to the probes from each of the arrays.
- http://www.cmis.csiro.au/IAP/Spot/spotmanual.htm; Buckley, M.J., CSIRO Mathematical and Information Sciences, user's guide for Spot microarray image analysis software.
- http://genome.ucsc.edu.; UCSC Genome Browser, used to identify splice variants (see Table 2).