Toward Rigorous Comprehension of Biological Complexity: Modeling, Execution, and Visualization of Thymic T-Cell Maturation (original) (raw)
Abstract
One of the problems biologists face is a data set too large to comprehend in full. Experimenters generate data at an ever-growing pace, each from their own niche of interest. Current theories are each able, at best, to capture and model only a small part of the data. We aim to develop a general approach to modeling that will help broaden biological understanding. T-cell maturation in the thymus is a telling example of the accumulation of experimental data into a large disconnected data set. The thymus is responsible for the maturation of stem cells into mature T cells, and its complexity divides research into different fields, for example, cell migration, cell differentiation, histology, electron microscopy, biochemistry, molecular biology, and more. Each field forms its own viewpoint and its own set of data. In this study we present the results of a comprehensive integration of large parts of this data set. The integration is performed in a two-tiered visual manner. First, we use the visual language of Statecharts, which makes specification precise, legible, and executable on computers. We then set up a moving graphical interface that dynamically animates the cells, their receptors, the different gradients, and the interactions that constitute thymic maturation. This interface also provides a means for interacting with the simulation.
What Do Biologists Try to Understand?
Biologists aim at understanding biological systems. Motivation varies from a desire to cure disease to pure fascination with living systems. The mark of biological systems is their complexity. Physicists have been the pioneers in trying to understand nature by reducing physical systems to component parts, which they analyze in detail. The biological equivalent has been to reduce complex organisms to their component cells and molecules and to analyze their behavior (Efroni and Cohen 2002, 2003). Biologists have taken up this challenge, and are in the process of cataloging the component parts of organisms at various scales. However, biological systems seem to resist this “understanding through reduction” for the following two reasons: (1) living systems are more complex than physical systems, and (2) in dissecting the molecular data, we remain far away from understanding the integrated living system.
Many groups have used mathematical tools to gain a better understanding of immunological data (for review, see Hood et al. 1980; Mehr et al. 1997, 1998; Gett and Hodgkin 2000; Hershberg et al. 2001; Bergmann et al. 2002; De Boer et al. 2003; Kesmir and De Boer 2003; Louzoun et al. 2003). The approach of these groups, however, differs fundamentally from ours in ways that will become apparent as we progress. Some of the differences have to do with our use of object-oriented visual specifications, and extensive run-time experimentation and visualization.
In this study, we use specific analytical data to construct an integrated dynamic representation. We carry out the integration via two interwoven facets. The first calls for specifying the data set in a way that makes it amenable to execution on a computer. The second generates an embodiment of the execution, representing the objects that are explicitly specified in the first facet, cells, and molecules. The end result is a moving visual simulation of the biological process—intuitive, visual, and interactive.
To form the first facet, a detailed description of the relevant objects is prepared. The task of collecting the data and translating it into a well-defined, executable specification is complex in itself. Scientific papers—the sources of the data—provide the data set in text, tables, and figures that are difficult to translate into other media. The language spoken in biological papers is usually comprehensible only to the specific field of research. Our goal here is to translate this data set into a generic and usable medium, which we refer to as the specification (or sometimes as the set of specifications).
The specifications derived from the actual data are used as instructions that guide the simulation. The cellular and molecular agents comprising the system refer, as it were, to these instructions to know how to respond to stimuli. The stimuli may be interactions with other cells, interactions with other molecules, or various internal events, such as the passage of time.
The task of specifying such a large data set needs its own special tools (for review, see Meier-Schellersheim 1999). Without such tools, it is difficult to control the immense set of data. The tool we use for specification (and, as we will show later, also for integration) is the language of Statecharts (Harel 1987), a visual formalism invented to aid in the design of complex man-made reactive systems, and later proposed as a viable tool for specifying biological systems in (N. Kam, I.R. Cohen, and D. Harel, in prep.). Below, we shall discuss the reasons for using this particular language. We begin here by detailing the problems mentioned above, as they arise in our model biological system—the thymus.
The Thymus as an Example of Disjoint Research
Stem cells arrive at the thymus from the bone marrow, and the developing T cells go through a series of interactions in different locations inside the thymus. The processes that a single cell goes through take about 2 wk (Anderson and Jenkinson 2001), during which time, the cell may proliferate into 106 offspring cells (Egerton et al. 1990). The thymic environment is divided anatomically into lobes and lobules, and the lobules are further divided into the areas of the cortex and the medulla. Because the thymic output of mature T cells is the basis of the immunological repertoire, the physiological function of the thymus is relevant to the study of many diseases, specifically AIDS and autoimmune diseases (Holoshitz et al. 1985; Cohen 2000; Douek et al. 2001, 2002).
Different agents constitute the thymus; epithelial cells form a mesh throughout the organ and interact with developing T cells to activate and regulate many of the processes needed for their maturation (Anderson and Jenkinson 2001; Germain 2002). Epithelial cells are separated into different subtypes by molecular markers or by anatomical location (Von Gaudecker et al. 1997). Macrophages perform mainly housekeeping tasks to clear the thymus of dead cells (Platt et al. 1996). Cytokines are the molecules responsible for signaling between the cells (Khaled and Durum 2002). Chemokines are molecules that signal cell movement along gradients (Norment and Bevan 2000; Zlotnik and Yoshie 2000). Short segments of proteins, called peptides, combine with other molecules (major histocompatibility molecules, MHC) to induce different T-cell selection events (e.g., Nanda and Sercarz 1995; Yasutomo et al. 2000). Thymocytes (T cells in the thymus) express many different surface molecules that serve as interactions with other cells and molecules. Epithelial cells, macrophages, cytokines, chemokines, peptides, thymocytes, and cell markers are all further divided into dozens of subgroups, which we need not detail here.
The thymic environment, loaded with these different objects, presents a challenge to many researchers from different fields who have detailed knowledge of some of its parts, but yet wish to comprehend the whole. Consider three scales of analysis—molecules, cells, and the whole organ.
Molecules
The molecules most relevant for researchers of the thymus, as we have said, are chemokines, cytokines, and receptors on the cell surface. Specialists in cell migration, for example, study how chemokines cause cell migration. They measure chemokine expression levels in different areas of the thymus, on different cells of the thymic stroma, and record the responses of thymocytes during different stages of their development. Biophysicists study the interactions between chemokine receptors and their chemokine ligands at the atomic level. Other researchers study cytokines and their influences on events in thymic development. Cytokines are the main vehicle for signaling between cells, and, therefore, are important in almost every process. Other molecules allow thymocytes to bind to other cells and to the extra-cellular matrix (ECM).
Other fields of research look at these molecules in a different way. In microscopy, molecules are used as markers to distinguish between different cells under the microscope. Researchers in signal transduction look at the same molecules to see how they influence a cascade of events inside the cell.
Cells
The questions asked at the cellular level are as follows. Which cells respond to which stimuli? How many cells of each type are in each thymic area? How many cell types are in various areas? What are the events that will drive a cell toward one fate and not another? What stages does a cell go through during development? Where is the cell during different stages of development? What are the paths two cells follow when they interact? Which selection events are the most influential? How does mutation influence cell survival?
Organ
Researchers looking at the thymus as one whole often see the organ as a black box. Their questions include the following: What is the number of cells the thymus produces under specific conditions? How many cells enter the thymus every day/hour/minute? What are the effects of removing the thymus (thymectomy)? Why does the thymus diminish in size with age? What are the influences of diseases on the thymus, and what is the influence of the thymus on disease? Are there mathematical formulas that can recapture thymic output behavior?
However, the thymus is one whole. Disjointed research parcels the same molecules and cells into separate fields, and produces data that must be joined if we are to ever understand T-cell maturation in the whole organ. Currently, there is no way to integrate this broad spectrum of different types of data into one view that would be as coherent as the biological environment that produced them. The work we present here is aimed at such integration. We take the data generated by reductionist biology and integrate them into a specification model using Statecharts. We then execute the model. The results of the execution are used to drive an animated visual image of cells and molecules and their interactions. This type of representation is friendly to human minds, and yet, does not sacrifice mathematical precision. Moving cells and molecules are interactive with the thoughts of the user, and the format provides the user with tools to choose specific views and to mediate particular types of execution.
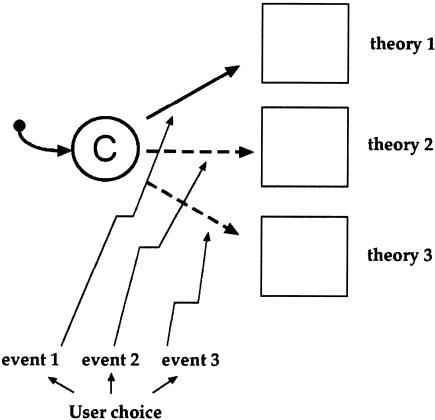
Moreover, we have designed representation to express different theories. Immunology, like other complex and incompletely characterized fields, uses theories to integrate both known and unknown information. Theories are proposed scenarios. Our model and simulation can accommodate different theories. Whenever an interaction takes place, the user (or the simulation itself) can choose one theory from a collection of available theories and instantiate that particular theory to its completion in the simulation. The instantiated theory then sends conclusions back to the simulation. The user can choose a particular theory either during run-time or during specification. The outcomes of various theories can be compared and contrasted.
RESULTS
Specifying the Thymus With Statecharts
States and Transitions as Descriptors of Cell Behavior
For specification and modeling, we use the language of Statecharts, a visual language invented by David Harel in 1984 (Harel 1987; Harel and Politi 1998) to assist in the development of the avionics system of a new aircraft. Statecharts has since become the subject of research for many groups (Wieringa 2003) as the main formalism used to specify the behavior of complex reactive systems in a variety of industries, and has been adopted as the central medium for describing behavior in the Unified Modeling Language (UML), a world standard for object-oriented specification and design of systems (Kobryn 1999).
Behavior in Statecharts is described using states and events that cause transitions between states. States may contain substates, thus enabling description at multiple levels, and zooming in and zooming out between levels. States may also be divided into orthogonal states, thus modeling concurrency, allowing the system to reside simultaneously in several different states. A cell, for example, may be described orthogonally as expressing several receptors, no receptors, or any combination of receptors at different stages of the cell cycle and in different anatomical compartments. Statecharts are rigorous and mathematically well defined, and are, therefore, amenable to execution by computers. Several tools have been built to support Statecharts-based modeling and execution, and to enable automatic translation from statecharts to machine code. We use a tool called Rhapsody (Harel and Gery 1997), commercially available from I-Logix, Inc.
It is not intuitively obvious that cells and molecules may be naturally described by states and transitions. In fact, there is no consensus on how one should describe cells. However, immunologists, whether they know it or not, do use states to describe cells. A cell is usually described by the collection of markers it expresses on its surface (Sant'Angelo et al. 1998). For example, a T cell is called double negative when neither of the CD4 and CD8 molecules is expressed. A human T cell is referred to as a memory cell when it expresses a molecule called CD45RO+ (Dutton et al. 1998) and as a suppressor cell when it coexpresses CD25 and CD4 without being activated (Cohen and Wekerle 1973; Mor et al. 1996; Elias et al. 1999; Coutinho et al. 2001; Shevach 2002). Immunologists call these molecules markers, but we refer to them, during specification, as orthogonal states of the cell. One may object to describing cells according to markers that are not chemically accurate descriptions. However, we use the notation, as it is the basis of most immunological reports and immunological terminology.
In Statecharts, transitions take the system from one state to another. In cell modeling, transitions are the result of biological processes or the result of user intervention. A biological process may be the result of an interaction between two cells, or between a cell and various molecules.
Dealing With a Large Data Set
Statecharts provide a controllable way to handle the enormous data set of cell behavior by providing us with the ability to specify separation into orthogonal states and by allowing transitions. For example, see Figure 1, which shows the statecharts of a single thymocyte. The thymocyte is a very complicated agent. To avoid clutter, the figure does not include all states and transitions, or all of the titles of the states it shows. By way of illustration, we have separated some of the orthogonal states and have indicated some of their sub-statecharts.
Figure 1.
A pseudo statechart of a thymocyte. The three-dimensional representation is our way of representing statecharts from different levels and showing their interrelationships.
Examples
Example 1: Modeling Thymocyte Movement
To demonstrate the way in which we convert data into specification, we shall follow the way thymocytes move in the thymus. Thymocytes receive signals from different cells in different locations. To make sure signals are received at the right time is actually to make sure that the right thymocyte is in the right place at the right time. The molecules responsible for directing cells along a gradient path are called chemokines. We focus on the role of the following four chemokines: CCL25 (TECK), CXCL12 (SDF), CCL22 (MDC), and CCL21 (SLC). Thymocytes search their environment for chemokines and move according to the chemokine gradient. We should therefore make sure that (1) the simulating gradient is correct, and (2) the thymocyte responds only to gradients it can currently interact with. To find out the right gradient, we survey the scientific literature to learn which chemokine is expressed where, and at what level. This information is available from different studies, ranging from papers whose subject is one specific chemokine and its expression in the thymus (Zaitseva et al. 2002), to papers dealing with one specific area in the thymus and the expression of different chemokines in that area (e.g., Chantry et al. 1999), to papers reviewing chemokine expression patterns in the thymus as a whole (e.g., Savino et al. 2002).
We integrate the chemokine data set to a four-dimensional lattice, in which each dimension stands for the concentration of one chemokine. Thymocytes first find out which of the gradients they should probe (we will explain how below), calculate the relevant gradient, and finally move.
To find which of the gradients a thymocyte may now probe, we use the notion, presented in the previous section, of cell types as cell states. In our model (as in immunology), we distinguish between cells according to surface markers. We ask which gradients are relevant at some specific stage. In other words, given a cell in a state characterized by the expression of certain markers and given a certain gradient, where will the cell move?
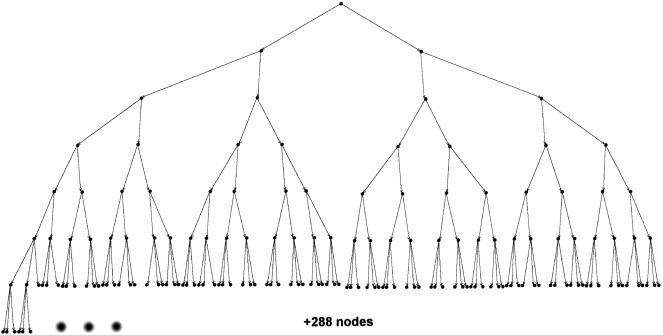
The scientific literature provides seven cell markers as relevant for gradient decisions. Five of them may be either expressed or unexpressed, and two of them have an intermediate level of expression termed “low”. The overall number of relevant states is therefore 25 × 32 = 288. At run time, a cell scans through these 288 states, finds the one it is in, and determines which chemokines it may respond to. Our job during specification is to go through these 288 states, find an equivalent in scientific papers and provide the biological meaning. During simulation, we use a decision tree to scan through the collection of possible states (Fig 2). Decisions (leafs of the last row) in the tree correspond to cell states. When the scan reaches a conclusion (a leaf), the simulation generates events that tell the cell to which chemokine gradients it may now respond.
Figure 2.
The 288 final nodes represent the final decisions of a thymocyte regarding which chemokine it may respond to. (The graph representing the tree was built with the DiGraph drawing algorithm described in Carmel et al. (2002).
Example 2: Modeling Epithelial Cells
Another example of specification is how we include epithelial cells in the model. Epithelial cells in the thymus are stationary; yet their behavior is reactive and changes continuously in response to various stimuli. The literature divides epithelial cells into many types. Because most of the work has been done using microscopy, the cell types are usually separated by their location and their size. To this microscopic division, we add temporal behavior, which is the expression of different chemokines and cytokines in response to different events. For example, medullary epithelial cells have shorter processes (arms) than other epithelial cell and are usually no longer than 30 µm in length. Medullary epithelial cells are considered the main elements in a process called negative selection, and, therefore, have been measured extensively for levels of expression of MHC class I and class II molecules.
We characterize epithelial cells as having not only a location, but also a structure. The structure is the cell processes (arms). As thymocytes and other cells move through the thymus, they interact with the processes of epithelial cells.
Specifying Interaction
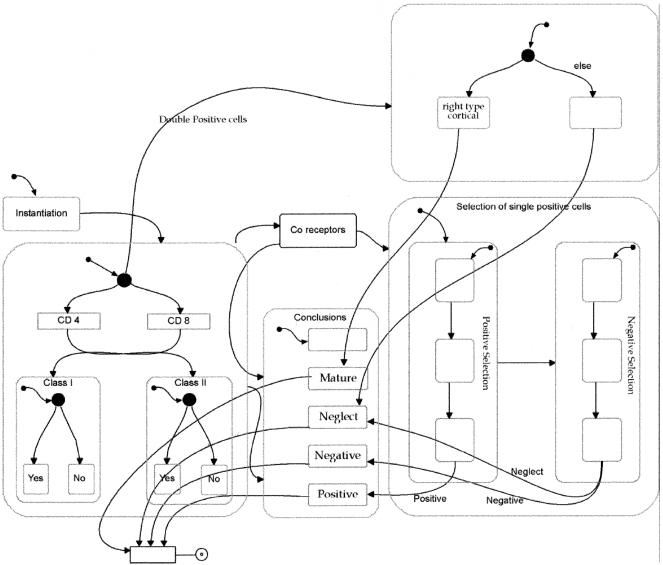
When two cells meet during run time, we need directions to tell us how their interaction should proceed. Researchers do not always know all the details of the interaction, and so they use different hypotheses to suggest possible outcomes of the interaction. We refer to the hypotheses and their suggested outcomes as theories, and outline them as objects with a behavior specified with Statecharts. Figure 3, for example, is the statechart of what we refer to as the classical epithelial cell—T-cell interaction. When we choose this theory, an instance of the theory is created every time a T cell and an epithelial cell meet.
Figure 3.
A theory of interactions between thymic epithelial cells and thymocytes presented as a statechart.
The statecharts of the instance are then executed, and according to different parameters, a conclusion of this interaction is reached. The conclusion may be the death of the T cell, instructions to express one or another marker, instructions to express cytokines, instructions to proliferate, and more. Eventually, the instance reaches the state marked with “T”, which means the instance is terminated and will receive no further references. When another interaction of the same kind takes place, another instance of the same kind will be instantiated. Notice that many instances may coexist as the result of many thymocyte-epithelial cell interactions occurring at the same time. According to a particular theory, a single epithelial cell may interact with many different T cells.
Using Statecharts to Communicate Theories
The diagrammatic nature of Statecharts makes them legible to scientists from different disciplines. To describe a theory with statecharts, we transform a description given in text and nonformal diagrams into a rigorous, diagrammatic language. The resulting description is easy to communicate. Figure 3, mentioned above, shows one such theory—the interaction of a T cell and an epithelial cell as described classically in textbooks (Janeway 2001).
Running Theories
By regarding theory as a separate component, we can choose to plug in or unplug a theory on demand. We build a collection of available theories and choose one of them. The choice of which theory should be instantiated may be made before we start the simulation. For example, we can decide that all interactions between thymocytes and cortical epithelial cells should follow one theory, whereas all other interactions follow a different theory. A choice of theory may also be made at run time, and the user can choose to switch between theories. The choice may also be made at run time by the simulation itself, when the right conditions develop. Theory, in our simulation, thus becomes interchangeable during the run. The simulation is only committed to the data, not to its interpretation.
The Front-End: An Interactive Animation
While the simulation runs, a front-end to its activities is generated and presented to the user. We have built the front-end as an interactive visual interface that embodies cells and molecules. The user can actually see what the cells and molecules are doing. The architecture to achieve this representation is described in the Methods section.
The General Setup
The representation is a large collection of Flash movie clips that are the embodiment of agents and their states as they appear in the simulation running in Rhapsody. While the simulation is generating events and is changing the properties of the interacting agents, the simulation sends information about these changes to generate the Flash movie. The animation is generated on the fly. The animation is neither an end result of the simulation, processed at post-run, nor a preprogrammed movie. It is a living image capturing the look and feel of the physical image of the simulated cells and molecules during run-time.
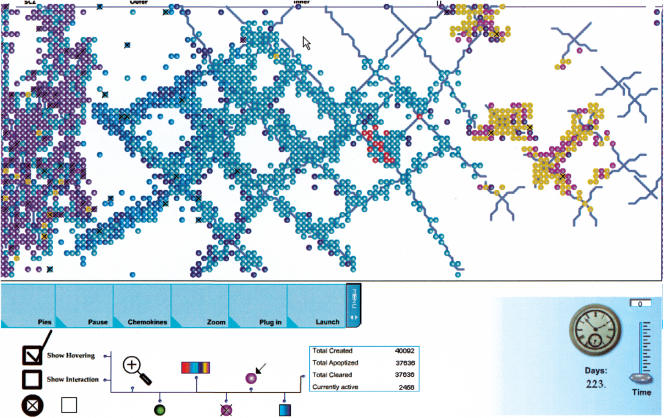
Movie M1 in the supporting online material shows a simulation during run-time. Briefly, we show an example of the interaction between the animation, the simulation, and the user in text and figures. Figure 4 gives a high-level view of a lobule at some point during execution. The figure serves only as an illustration to show what the front-end looks like. We briefly detail the parts mentioned in the figure. The buttons Pies, Pause, Chemokines, Zoom, Plug in, and Launch control the (accordingly) statistical representation of the data; pause the simulation; chemokine representation; different zooming in-and-out abilities; connection between the animation and simulation. The other buttons give different color codes relevant to the display, enable the user to trace the motion of specific cells, control the connection between the simulation and specific statistical tool (such as Matlab), give the user the ability to avoid clutter made by overlapping cells, give the user the ability to receive visual indication to interactions, and more. The clock shows how much biological time has gone by since we began. We use the term “biological” time to emphasize its difference from “chronological” time. The slide bar above the clock gives us the ability to compress time, and the caption next to the slide bar tells us by what degree. For example, if the caption shows the number 30, then 1 sec in biological time is transformed into 0.3 sec of chronological runtime. The small circles in Figure 4 are the visual representation of thymocytes. At the level of detail in the figure, it is not possible to show how the cells are different, especially in their surface markers, but also in other features we use to model their dynamic behavior. However, more than just an embodiment of the underlying code, the thymocytes serve as an interactive user interface. By clicking the cell surface, the user is presented with a menu allowing control over cell attributes, states, and destiny, and with tools to obtain information about the simulation, the user cannot perceive from the current view. We detail the interactive implement below.
Figure 4.
A snapshot of the simulation during run time.
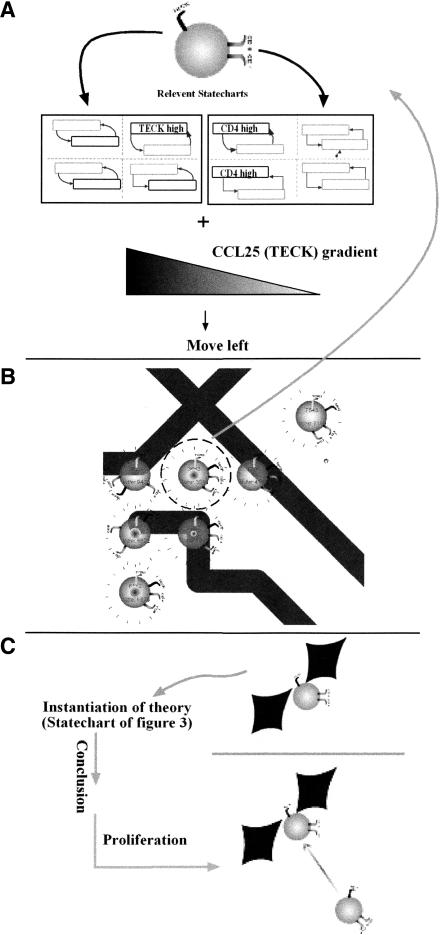
Two Examples
Figure 5 portrays in part B how one thymocyte moves, and in part C, how an interaction with an epithelial cell takes place. Figure 5A gives a snapshot of the running simulation. The figure shows collections of thymocytes around one epithelial cell in the animated user interface. It is important to emphasize that the image of the thymocytes is not a sketch made for the figure, but a screen capture of the running simulation.
Figure 5.
Decision making during simulation. The thyocyte surrounded by a circle in A decides where to migrate according to statecharts similar to the ones portrayed in B. The thymocyte in C, after making physical contact with an epithelial cell, instantiated the theory portrayed in Figure 3, concluding that it should proliferate.
In Figure 5, B and C, we show a sketch of two mechanisms that determine the behavior of the cells. In B, below the image of the thymocyte, we show parts of the statechart of the thymocyte. We show only two sub-statecharts corresponding to the three markers visible on the cell's surface, and not the full statechart that would look similar to Figure 1. The thymocyte currently expresses the receptors CD4 and CD8 (the immunological term is DP—double positive) and is responsive to the chemokine CCL25 (TECK). Contrary to the two markers for CD4 and CD8, which stand for real surface molecules with that name, the marker for CCL25 (TECK) does not signify a molecule, but signifies the ability of the thymocyte to migrate according to a gradient created by that specific chemokine. We use this notation because the experimenters have only limited knowledge of which receptors cause which movements. The available data experimenters provide is of the form “which T cell migrates according to which chemokine” (Kim et al. 1998; Campbell et al. 1999; Norment and Bevan 2000; Annunziato et al. 2001; Taylor et al. 2001; Savino et al. 2002). The sub-statecharts show how we represent receptors as orthogonal states. An expressed receptor will be in the state high, and an unexpressed receptor will be in the state low. On the left statechart, we see only one state in high. The state represents susceptibility to CCL25 (TECK) migration. On the right side, two receptors are in high—CD4 and CD8.
To be able to move, the thymocyte represented in the figure (as all other cells) continuously samples its environment. When the thymocyte finds a relevant chemokine gradient—a CCL25 (TECK) gradient—it calculates the gradient difference across its surface. Cell movement is directed according to this calculation. In this example, the conclusion is for the thymocyte to move left.
Figure 5C portrays a different mode of operation. The lower part of Figure 5A shows a thymocyte next to part of the arm of an epithelial cell, represented as the two adjacent red diamonds. The thymocyte has just migrated from the right and touched the epithelial cell to its left. When the thymocyte and the epithelial cell meet, they instantiate the behavior of the statechart described in the previous section. It is the same statechart we used in Figure 3. The conclusion of this specific interaction is the result of several checks made during the execution of the statechart, which checks the states, the thymocyte, the attributes of the thymocyte, and the properties of the epithelial cell, and finally comes up with the conclusion that, in this case, the specific thymocyte should now proliferate. Proliferation will result in the creation of other thymocytes bearing the same markers and having the same attributes as the parent cell. The proliferation updates the Flash movie. When a new thymocyte is created in the movie, an arrow to designate its ancestor appears and then vanishes.
The simulation handles many such events during run-time. Thymocytes continually move around in the simulated thymus, continuously check their environment for stimuli, respond to the stimuli, proliferate, mature, die, change their receptors, secrete cytokines, and interact with other cells. All of this is displayed at run-time on the user interface and in animated state charts generated by Rhapsody. Because every agent in the simulation is, in effect, an instance in Rhapsody, the user may choose to focus on an animated statechart of the agent. Animated statecharts are useful when we wish to study, in detail, events and switches in states during simulation. We may, for example, wish to follow the details of the interaction that resulted in migration toward the medulla. Because Rhapsody provides a step-by-step mode, we can interrupt the flow of the simulation at any time and continue one step at a time, while paying attention to relevant attributes and following any switches in states the cells go through. We follow choices made by theory instances and watch them arrive at decisions. This course of action may be referred to as “debugging” the simulated biological process. We debug at two levels. First, we watch the visual embodiment of the simulation as it develops in the animated representation. We look for emerging patterns, for dead-end paths, for undefined observables, and for mistakes. To carefully scrutinize parts and time bites, we use the power of animated statecharts and progress step-wise. This allows us to look at every agent as one reactive system, and to handle the flood of incoming/outgoing events in a controllable way.
Interactivity
Both the visual user interface and the underlying executed animated statecharts allow the user to manipulate the simulation and to retrieve data. This is done in two separate ways. We shall first explain interactions via the visual user interface, and then explain how the user directly manipulates statecharts.
Interactions Via User Interface
As we explained above, the front-end of the simulation is composed of a collection of movie clips. Each of the movie clips is, in fact, an interactive menu that allows the user to send data to the running simulation. Because the sent data is, in fact, an XML object (see Methods), we are not limited in its contents. We perceive available operations as belonging to one of two kinds, data manipulation or data request.
Data Manipulation
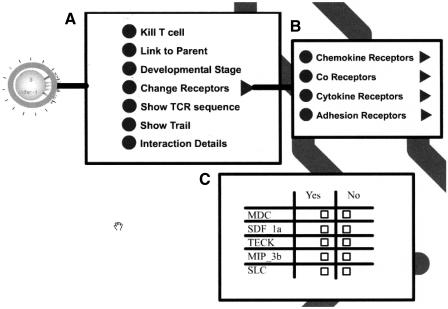
Every object in the animation is also a clickable menu. We demonstrate data manipulation and data request upon clicking the animated thymocyte. In Figure 6, you can see the menu that opens when the user clicks a thymocyte. The menu item “kill T cell” serves as an example of data manipulation. When the user clicks this item, the underlying executing simulation receives notification that it should now tell this specific T cell to perform apoptosis (programmed death). The results of apoptosis are performed in the simulation itself. When the results are processed, the animation will receive the instruction from the simulation to now delete the thymocyte from current view (and to perform any other representation tasks needed).
Figure 6.
An example of menus that open in response to clicking a thymocyte.
The submenu Change Receptors opens into four submenus that control the cell's receptors (Fig. 6b). The figure shows the submenu that opens the menu item Chemokine Receptors (Fig. 6c). By clicking any element in the checkbox table, the user can change the ability of the cell to migrate to any of the chemokines. For example, upon clicking the checkbox in MDC/Yes, the animation sends an event to the simulation. The simulation will then do two things; it will direct the cell that it may now migrate according to CCL22 (MDC), and it informs the animation that the thymocyte should now indicate that it is susceptible to CCL22 (MDC) [by showing the CCL22 (MDC) indicator]. The user thus manipulates the simulation exactly in the same way data manipulate the simulation. Data manipulation events originating from the user are no different, as far as the simulation is concerned, from events that stem from data specification.
Data Retrieval
In contrast to data manipulation, data retrieval events do not direct or drive the simulation process. The menu items Link to Parent, Developmental Stage, and Show TCR sequence of Figure 6A are examples of retrieval events.
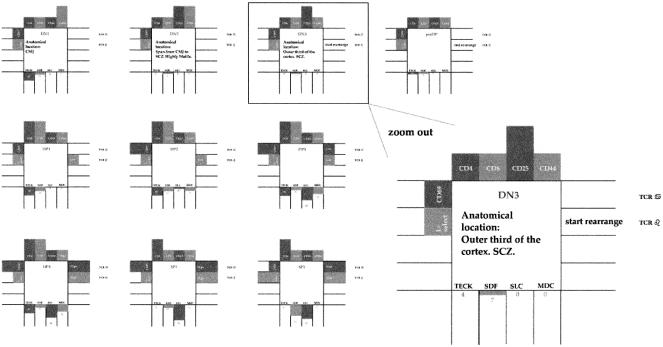
The menu item Developmental Stage opens the diagram shown in Figure 7 that describes the path of development that thymocytes go through in the thymus as current research sees it. The path, as we discussed above, is in fact, a description of which markers are now on the thymocyte surface. The diagram that opens in response to the click indicates graphically which developmental stage the thymocyte is currently in. As we explain in the Methods section, we make available the publications that constitute the factual basis for this diagram. By clicking the diagram, the relevant paper is retrieved.
Figure 7.
A visual representation of the developmental stages thymocytes go through. The representation also shows, together with conventional markers, the migratory abilities of each developmental stage. During run-time, the user may click on an animated thymocyte to retrieve the representation shown in the figure, with the appropriate stage highlighted. Further, each of the marked receptors and molecules serves as a button. A click on this button retrieves the scientific paper that is the source for the inclusion of the receptor, molecule, or any scientific detail.
The menu item Show TCR sequence simply gives the amino acid sequence of the T-cell antigen receptor (TCR). Currently, we are in the process of providing the user with more data retrieval options (see Discussion).
Direct Interactions With the Statecharts
While the simulation is running, the user can interact with the underlying statecharts directly, with tools available from Rhapsody, without using the interface, by injecting events. In other words, during run-time, the user may choose a specific event to be performed immediately. The run continues, the chosen event is inserted, and it effects the simulation directly.
Figure 8 provides an example. The pseudo-statechart in the figure gives part of the statechart of an epithelial cell. As explained above, the user can choose from any of the three theories in the figure. A choice is made during run time, when the user finds the needed instance of an epithelial cell in Rhapsody, decides the theory he or she would like the cell to implement, and injects the appropriate event—1, 2, or 3.
Figure 8.
User intervention directly influencing statecharts.
This is similar to using a switch mechanism to direct a train to a railroad track of choice.
Tracing Back the Data
We have made an effort to set things up so that the data we use—scientific papers, tables, figures, and diagrams—are available to the user during run time.
Figure 7 demonstrates this as follows: The figure is a representation of several stages a thymocyte goes through during development. The figure is, in fact, a compilation of the data found in Ritter and Crispe (1991), Ritter and Crispe (1992), Penit et al. (1995) Tourigny et al. (1997), Chantry et al. (1999), Youn et al. (1999), Bleul and Boehm (2000), Norment and Bevan (2000), Annunziato et al. (2001), Lind et al. (2001), Hernandez-Lopez et al. (2002), and in it we use the same taxonomy used for cell states and for cell markers, with the exception that we also indicate a cell's susceptibility to chemokines as markers with assigned probability. For example, the first stage in the figure—DN1—represents a population of cells, of which 40% migrate to CCL25 (TECK), 12% migrate to CXCL12 (SDF), etc. We use this figure when we want to retrieve the data and implement it in a manageable way into specification.
This figure can be used at run time. When a user wants to examine the reasons for a cell's movement, he or she may click on the specific cell and choose the menu item Developmental Stage. This action opens the same diagram we use for specification—Figure 7—only with an indication to the current state of the cell (the current state is marked with a rectangle around the appropriate stage). Further, if the user wishes to retrieve a paper that serves as the basis for any of the data represented in the figure, he or she need only click the specific item in the figure and a window containing the paper opens up.
We also make tracing of data available in the statechart specification itself. Every state and every transition in a statechart has a field called description, to which we have attached references to relevant papers. In this way, a user who chooses to view the running simulation through its animated statecharts, can find the reasons behind some of the choices. The references are especially useful when we specify theories.
As we described previously, a theory object is closely related to a scientific paper or to a group of scientific papers representing an hypothesis. By directly linking the statechart representing of the theory and the paper describing the hypothesis, we fashion a trace not only to the data, but also to its interpretation.
METHODS
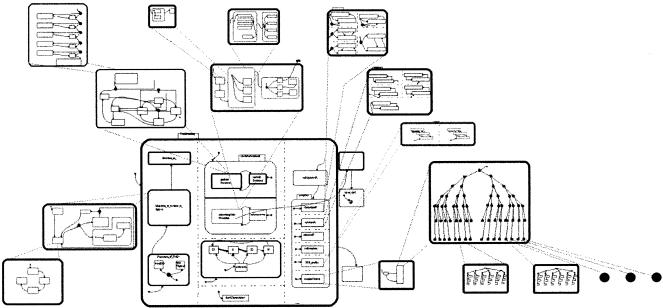
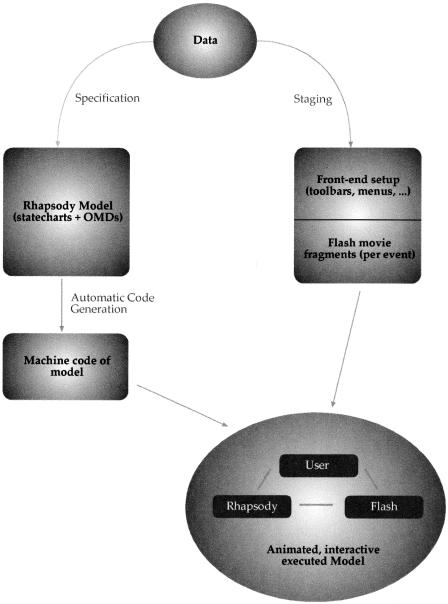
The scheme we use is represented in Figure 9. For a detailed description, see Harel et al. (2003). To draw statecharts and object model diagrams (OMD), we use a code generation and implementation tool called Rhapsody from I-Logix (for review, see Harel and Gery 1997). After generating and compiling the code, we can run the application with Rhapsody animating the statecharts.
Figure 9.
The software setup that enables the modeling, simulation, and interactive animation.
Animation of the visual user interface is done with Flash. For this, we have built a collection of Flash movie clips that represent the cells and molecules. On top of this collection, we encode a set of instructions that tells the Flash movie how to respond to events from the simulation. For example, expressing a receptor would start, in the Flash movie, a cascade that (1) finds the movie that represents the thymocytes, (2) finds the movie that represents the receptor, (3) attaches the receptor movie in the right place relative to the thymocyte movie, and (4) starts playing the receptor movie.
The connection between the simulation and the animation is done using TCP/IP channels. The events themselves are XML objects. Flash can receive XML messages through an object, available in Flash, called an XML socket. In the simulation, we implement a server that channels communication to the proper TCP/IP socket and receives XML messages sent back from the animation, by mouse clicks made by the user. Events on both sides are also written in XML. On the simulation side, we parse the incoming XML objects with common tools for XML parsing. The Flash movie parses XML objects with tools available in Flash. As XML objects are very useful for communicating any data structure, we are practically unlimited in our ability to convey instructions between the two arms of the run-time environment. In time, we expect to add more power to this communication.
DISCUSSION
Future Work
Other than obvious improvements to our simulation, model, animation, and user interface (better, faster implementation; better architecture; improved convenience of the user interface; capturing a larger part of the data set; implementing more theories, etc.), we believe future work should go in two directions, the lower scale and the upper scale.
The Lower and Higher Scale
In the study reported upon here, we artificially decrease complexity to enable modeling. We work in two dimensions—we take thymocytes and macrophages to be of fixed size and shape, and we represent the multiple copies of receptors on a cell's surface (the way immunologists do) as one receptor; we work on a lattice with some predefined resolution. The assumptions can be treated differently if we switch to a lower scale—the molecular scale. On the molecular scale, cells are represented as actual collections of molecules, and we no longer transform molecular collections into cells, but simulate molecular collections. We do not choose between theories, but simulate interacting cells as their molecules bind and interact.
However, the molecular scale is currently impractical. Not enough data is available about interactions at the molecular level. The complexity at this level would result in an effort directed only at a single cell, and would make higher levels of perspective—a cell population and an organ—practically impossible to achieve. There have been remarkable efforts to simulate single cells at the molecular level (Tomita 2001; Bartol and Stiles 2002). For this kind of simulation, the groups must use supercomputing power. Therefore, for the time being, we cannot even attempt to go from the molecular description level of one cell to the level of cell populations.
A higher scale does not require a change in specification and implementation, but needs a different perspective to look at information generated by the simulation. While the simulation runs, cells and molecules are generated and change their properties. A lot of information is available about these cells, their types, their attributes, their locations, their history, the history of their interactions, their relations to other cells, etc.
We believe that new ways to look at data must be found, and new tools to support them must be built as information visualization itself changes the questions asked. We are in the process of building such tools. Population size tools, unlike molecular level tools, do not need special machinery, as special algorithms are made available using current computer architecture. Population level analysis is a relevant scale when we look at most functions of the immune system. The immune system eradicates pathogens by changing the ratios of cell numbers in different clones; the immune system maintains homeostasis by controlling population ratios; pharmaceutical drugs usually work on specific populations of cells defined as bearing the same markers. The population view is the natural view for immunologists.
Ex Vivo Experimentation
The work described in this study is work in progress, and it remains to be applied to direct experimentation and to theoretical comparisons. We are in the process of fine-tuning our tools to make them available for such implementation and to study defined immunological phenomena.
Conclusions
Biological understanding is specific to the problem at hand and to the scale in question. We think we understand a biological system when we can make predictions about it, when we can utilize it, or when we can rephrase its meaning (I.R. Cohen, in prep). Much of the work done so far in systems biology has been directed at understanding the genome. This work has generated its own terminology. In this study, we use the words data, information, modeling, simulation, hypothesis, and even systems with meanings that may be different from those used in genomic bioinformatics. However, the problem of understanding is the issue, not terminology.
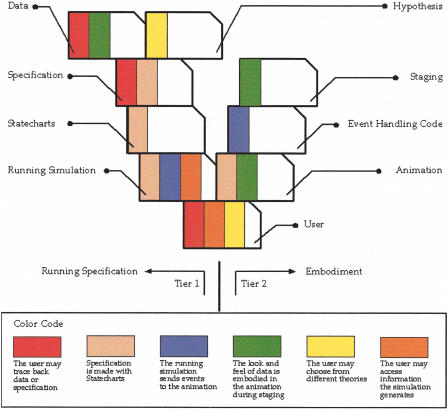
This study presents a two-tier strategy for comprehending the biological complexity of the thymus. The combination of these two tiers makes the effort manageable, executable, and comprehensible.
Tier 1 may be seen as the mathematical modeling of available data to model the thymus with tools invented in computer science for system analysis and system design. The tools make the analysis legible and mathematically valid with the help of the visual language of Statecharts. The mathematical rigor of the model makes it amenable to execution on a computer as a running simulation. We use this simulation to perform experimentation—thought experiments if you will—with an existing data set. With the proper configuration, we provide the added ability to switch between different theories proposed to explain the data set.
The end result of Tier 1 is a running simulation; see Figure 10. Although the simulation is of value in its own right—products of the simulation can be analyzed at run time or post-run—our goal is a lucid representation of the information generated by the simulation. This representation is the end product of Tier 2.
Figure 10.
A general view of the methodology used in this work. The left side displays the procedure of turning scientific data into computer-legible specification. The right side displays the procedure of building self-constructed animation through building animation components and their instructional combinations.
Tier 2 is the embodiment of cells and molecules. Different embodiments of cells and molecules are at the heart of biological explanations and biological understanding. These embodiments are usually sketches, movies, visual explanations, or textbook diagrams. There are even traditional conventions for the diagrammatic representation of cells—they should be round. The diagrammatic representation of molecules is usually specific for a particular field of study. The explanatory power of the visual is the motivation for building Tier 2. Here, however, our front-end departs from traditional biological representations, which are staged, either by being static or by being preplanned. The front-end result of our Tier 2 is not staged. The running simulation continuously generates the representation. This front-end thus maintains its explanatory power while adhering to the specified data as it is supplied in Tier 1.
The agents that are the basis for specification in Tier 1 are imaged in Tier 2. The cells and molecules become animated, interactive movies. This interactivity allows manipulation and representation of the data that generated the simulation and the data that is generated by the simulation. We thus supply a new link between the scientists who use the simulation and the scientists who provide the data. We also supply new links within the data set itself. Data that arrive from different papers and from different fields are recombined to form the whole organ or organism that generates the data. The data recombine because specification necessitates such integration. The detailed specification of one cell is the fused mass of data.
Here, we show a methodology and an implementation for incorporating large amounts of data regarding one biologically interesting environment. In addition to recording the cells and molecules comprising the system and capturing the dynamics of their interactions, a most valuable contribution of such an approach will be the ability to make prediction and carry out a pilot experiment in silico. Such experimentation will challenge the value of our approach. Preliminary studies suggest that it is possible to perform experiments in out system.
It now seems that we can process the information needed to get some understanding of cell populations. For example, we have been able to explore questions such as the percentage of all thymocytes bearing particular markers that are responsive to CCL25 (TECK). We can also determine how many thymocytes stem from one progenitor and how many thymocytes die from neglect or from negative selection. We can identify the T cells that encounter a specific macrophage throughout its history. Any immunologist can come up with many more interesting global questions. Such in silico experimentation will be the subject of future publications.
Acknowledgments
This work was supported by grants from the Minerva Foundation and by the Robert-Koch Minerva Center for the Study of Autoimmune Diseases.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1215303.
[Supplemental material is available online at www.genome.org and at www.wisdom.weizmann.ac.il/sol/sysbio2002/.\]
References
- Anderson, G. and Jenkinson, E.J. 2001. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 1**:** 31-40. [DOI] [PubMed] [Google Scholar]
- Annunziato, F., Romagnani, P., Cosmi, L., Lazzeri, E., and Romagnani, S. 2001. Chemokines and lymphopoiesis in human thymus. Trends Immunol. 22**:** 277-281. [DOI] [PubMed] [Google Scholar]
- Bergmann, C., van Hemmen, J.L., and Segel, L.A. 2002. How instruction and feedback can select the appropriate T helper response. Bull. Math. Biol. 64**:** 425-446. [DOI] [PubMed] [Google Scholar]
- Bleul, C.C. and Boehm, T. 2000. Chemokines define distinct microenvironments in the developing thymus. Eur. J. Immunol. 30**:** 3371-3379. [DOI] [PubMed] [Google Scholar]
- Campbell, J.J., Pan, J., and Butcher, E.C. 1999. Cutting edge: Developmental switches in chemokine responses during T cell maturation. J. Immunol. 163**:** 2353-2357. [PubMed] [Google Scholar]
- Carmel, L., Harel, D., and Koren, Y. 2002. Drawing directed graphs using one-dimensional optimization. In Comp. Sci. 2528: Proc. Graph Drawing 2002, pp. 193-206.
- Chantry, D., Romagnani, P., Raport, C.J., Wood, C.L., Epp, A., Romagnani, S., and Gray, P.W. 1999. Macrophage-derived chemokine is localized to thymic medullary epithelial cells and is a chemoattractant for CD3(+), CD4(+), CD8(low) thymocytes. Blood 94**:** 1890-1898. [PubMed] [Google Scholar]
- Cohen, I.R. 2000. Tending adam's garden: Evolving the cognitive immune self. Academic Press, London, UK.
- Cohen, I.R. and Wekerle, H. 1973. Regulation of T-lymphocyte autosensitization. Transplant Proc. 5**:** 83-85. [PubMed] [Google Scholar]
- Coutinho, A., Hori, S., Carvalho, T., Caramalho, I., and Demengeot, J. 2001. Regulatory T cells: The physiology of autoreactivity in dominant tolerance and “quality control” of immune responses. Immunol. Rev. 182**:** 89-98. [DOI] [PubMed] [Google Scholar]
- De Boer, R.J., Mohri, H., Ho, D.D., and Perelson, A.S. 2003. Turnover rates of B cells, T cells, and NK cells in simian immunodeficiency virus-infected and uninfected rhesus macaques. J. Immunol. 170**:** 2479-2487. [DOI] [PubMed] [Google Scholar]
- Douek, D.C., Betts, M.R., Hill, B.J., Little, S.J., Lempicki, R., Metcalf, J.A., Casazza, J., Yoder, C., Adelsberger, J.W., Stevens, R.A., et al. 2001. Evidence for increased T cell turnover and decreased thymic output in HIV infection. J. Immunol. 167**:** 6663-6668. [DOI] [PubMed] [Google Scholar]
- Douek, D.C., Brenchley, J.M., Betts, M.R., Ambrozak, D.R., Hill, B.J., Okamoto, Y., Casazza, J.P., Kuruppu, J., Kunstman, K., Wolinsky, S., et al. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417**:** 95-98. [DOI] [PubMed] [Google Scholar]
- Dutton, R.W., Bradley, L.M., and Swain, S.L. 1998. T cell memory. Annu. Rev. Immunol. 16**:** 201-223. [DOI] [PubMed] [Google Scholar]
- Efroni, S. and Cohen, I.R. 2002. Simplicity belies a complex system: A response to the minimal model of immunity of Langman and Cohn. Cell. Immunol. 216**:** 23-30. [DOI] [PubMed] [Google Scholar]
- ____. 2003. The heuristics of biologic theory: The case of self-nonself discrimination. Cell. Immunol. 223**:** 87-89. [DOI] [PubMed] [Google Scholar]
- Egerton, M., Scollay, R., and Shortman, K. 1990. Kinetics of mature T-cell development in the thymus. Proc. Natl. Acad. Sci. USA 87**:** 2579-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias, D., Tikochinski, Y., Frankel, G., and Cohen, I.R. 1999. Regulation of NOD mouse autoimmune diabetes by T-cells that recognize a TCR CDR3 peptide. Int. Immunol. 11**:** 957-966. [DOI] [PubMed] [Google Scholar]
- Germain, R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2**:** 309-322. [DOI] [PubMed] [Google Scholar]
- Gett, A.V. and Hodgkin, P.D. 2000. A cellular calculus for signal integration by T-cells. Nat. Immunol. 1**:** 239-244. [DOI] [PubMed] [Google Scholar]
- Harel, D. 1987. Statecharts: A visual formalism for complex systems. Sci. Comput. Programm. 8**:** 231-274. [Google Scholar]
- Harel, D. and Gery, E. 1997. Executable object modeling with statecharts. IEEE Comput. 30**:** 31-42. [Google Scholar]
- Harel, D. and Politi, M. 1998. Modeling reactive systems with statecharts: The statemate approach. McGraw-Hill, New York.
- Harel, D., Efroni, S., and Cohen, I.R. 2003. Reactive animation. Lecture Notes in Computer Science (in press).
- Hernandez-Lopez, C., Varas, A., Sacedon, R., Jimenez, E., Munoz, J.J., Zapata, A.G., and Vicente, A. 2002. Stromal cell-derived factor 1/CXCR4 signaling is critical for early human T-cell development. Blood 99**:** 546-554. [DOI] [PubMed] [Google Scholar]
- Hershberg, U., Louzoun, Y., Atlan, H., and Solomon, S. 2001. HIV time hierarchy: Winning the war while losing all the battles. Physica A 289**:** 178-190. [Google Scholar]
- Holoshitz, J., Matitiau, A., and Cohen, I.R. 1985. Role of the thymus in induction and transfer of vaccination against adjuvant arthritis with a T lymphocyte line in rats. J. Clin. Invest. 75**:** 472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, J.M., Huang, H.V., and Hood, L. 1980. A computer simulation of evolutionary forces controlling the size of a multigene family. J. Mol. Evol. 15**:** 181-196. [DOI] [PubMed] [Google Scholar]
- Janeway, C. 2001. Immunobiology: The immune system in health and disease. Garland Pub., New York.
- Kesmir, C. and De Boer, R.J. 2003. Clonal exhaustion as a result of immune deviation. Bull. Math. Biol. 65**:** 359-374. [DOI] [PubMed] [Google Scholar]
- Khaled, A.R. and Durum, S.K. 2002. The role of cytokines in lymphocyte homeostasis. Biotechniques Suppl: 40-45. [PubMed]
- Kim, C.H., Pelus, L.M., White, J.R., and Broxmeyer, H.E. 1998. Differential chemotactic behavior of developing T-cells in response to thymic chemokines. Blood 91**:** 4434-4443. [PubMed] [Google Scholar]
- Kobryn, C. 1999. UML 2001: A standardization odyssey. Comm. of the ACM 42**:** 29-37. [Google Scholar]
- Lind, E.F., Prockop, S.E., Porritt, H.E., and Petrie, H.T. 2001. Mapping precursor movement through the postnatal thymus reveals specific microenvironments supporting defined stages of early lymphoid development. J. Exp. Med. 194**:** 127-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louzoun, Y., Weigert, M., and Bhanot, G. 2003. Dynamical analysis of a degenerate primary and secondary humoral immune response. Bull. Math. Biol. 65**:** 535-545. [DOI] [PubMed] [Google Scholar]
- Mehr, R., Perelson, A.S., Fridkis-Hareli, M., and Globerson, A. 1997. Regulatory feedback pathways in the thymus. Immunol. Today 18**:** 581-585. [DOI] [PubMed] [Google Scholar]
- Mehr, R., Perelson, A.S., Sharp, A., Segel, L., and Globerson, A. 1998. MHC-linked syngeneic developmental preference in thymic lobes colonized with bone marrow cells: A mathematical model. Dev. Immunol. 5**:** 303-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Schellersheim, M. 1999. “SIMMUNE, a tool for simulating and analyzing immune system behavior.” Dissertation, University of Hamburg, Hamburg, Germany.
- Mor, F., Reizis, B., Cohen, I.R., and Steinman, L. 1996. IL-2 and TNF receptors as targets of regulatory T-T interactions: Isolation and characterization of cytokine receptor-reactive T-cell lines in the Lewis rat. J. Immunol. 157**:** 4855-4861. [PubMed] [Google Scholar]
- Nanda, N.K. and Sercarz, E.E. 1995. The positively selected T-cell repertoire: Is it exclusively restricted to the selecting MHC? Int. Immunol. 7**:** 353-358. [DOI] [PubMed] [Google Scholar]
- Norment, A.M. and Bevan, M.J. 2000. Role of chemokines in thymocyte development. Semin. Immunol. 12**:** 445-455. [DOI] [PubMed] [Google Scholar]
- Penit, C., Lucas, B., and Vasseur, F. 1995. Cell expansion and growth arrest phases during the transition from precursor (CD4-8-) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J. Immunol. 154**:** 5103-5113. [PubMed] [Google Scholar]
- Platt, N., Suzuki, H., Kurihara, Y., Kodama, T., and Gordon, S. 1996. Role for the class A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes in vitro. Proc. Natl. Acad. Sci. 93**:** 12456-12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter, M.A. and Crispe, I.N. 1991. The thymus. IRL Press at Oxford University Press, Oxford, UK.
- Ritter, M.A. and Crispe, T.N. 1992. The Thymus. Oxford University Press, New York.
- Sant'Angelo, D.B., Lucas, B., Waterbury, P.G., Cohen, B., Brabb, T., Goverman, J., Germain, R.N., and Janeway, C.A.J. 1998. A molecular map of T-cell development. Immunity 9**:** 179-186. [DOI] [PubMed] [Google Scholar]
- Savino, W., Mendes-da-Cruz, D.A., Silva, J.S., Dardenne, M., and Cotta-de-Almeida, V. 2002. Intrathymic T-cell migration: a combinatorial interplay of extracellular matrix and chemokines? Trends Immunol. 23**:** 305-313. [DOI] [PubMed] [Google Scholar]
- Shevach, E.M. 2002. CD4+ CD25+ suppressor T-cells: More questions than answers. Nat. Rev. Immunol. 2**:** 389-400. [DOI] [PubMed] [Google Scholar]
- Taylor, J.R.J., Kimbrell, K.C., Scoggins, R., Delaney, M., Wu, L., and Camerini, D. 2001. Expression and function of chemokine receptors on human thymocytes: Implications for infection by human immunodeficiency virus type 1. J. Virol. 75**:** 8752-8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita, M. 2001. Whole-cell simulation: A grand challenge of the 21st century. Trends Biotechnol. 19**:** 205-210. [DOI] [PubMed] [Google Scholar]
- Tourigny, M.R., Mazel, S., Burtrum, D.B., and Petrie, H.T. 1997. T-cell receptor (TCR)-β gene recombination: Dissociation from cell cycle regulation and developmental progression during T-cell ontogeny. J. Exp. Med. 185**:** 1549-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Gaudecker, B., Kendall, M.D., and Ritter, M.A. 1997. Immuno-electron microscopy of the thymic epithelial microenvironment. Microsc. Res. Tech. 38**:** 237-249. [DOI] [PubMed] [Google Scholar]
- Wieringa, R. 2003. Design methods for reactive systems: Yourdan, Statemate, and the UML. Morgan Kaufmann Publishers, Amsterdam, Boston, MA.
- Yasutomo, K., Lucas, B., and Germain, R.N. 2000. TCR signaling for initiation and completion of thymocyte positive selection has distinct requirements for ligand quality and presenting cell type. J. Immunol. 165**:** 3015-3022. [DOI] [PubMed] [Google Scholar]
- Youn, B.S., Kim, C.H., Smith, F.O., and Broxmeyer, H.E. 1999. TECK, an efficacious chemoattractant for human thymocytes, uses GPR-9-6/CCR9 as a specific receptor. Blood 94**:** 2533-2536. [PubMed] [Google Scholar]
- Zaitseva, M., Kawamura, T., Loomis, R., Goldstein, H., Blauvelt, A., and Golding, H. 2002. Stromal-derived factor 1 expression in the human thymus. J. Immunol. 168**:** 2609-2617. [DOI] [PubMed] [Google Scholar]
- Zlotnik, A. and Yoshie, O. 2000. Chemokines: A new classification system and their role in immunity. Immunity 12**:** 121-127. [DOI] [PubMed] [Google Scholar]