Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells (original) (raw)
. Author manuscript; available in PMC: 2014 Jul 4.
Published in final edited form as: Nat Chem. 2013 Nov 24;6(1):75–80. doi: 10.1038/nchem.1805
Following extensive evidence for the formation of four-stranded DNA G-quadruplex structures in vitro, DNA G-quadruplexes have been observed within human cells. Although chemically distinct, RNA can also fold in vitro into G-quadruplex structures that are highly stable because of the 2′-hydroxyl group. However, RNA G-quadruplexes have not yet been reported in cells. Here, we demonstrate the visualization of RNA G-quadruplex structures within the cytoplasm of human cells using a G-quadruplex structure-specific antibody. We also demonstrate that small molecules that bind to G-quadruplexes in vitro can trap endogenous RNA G-quadruplexes when applied to cells. Furthermore, a small molecule that exhibits a preference for RNA G-quadruplexes rather than DNA G-quadruplexes in biophysical experiments also shows the same selectivity within a cellular context. Our findings provide substantive evidence for RNA G-quadruplex formation in the human transcriptome, and corroborate the selectivity and application of stabilizing ligands that target G-quadruplexes within a cellular context.
In nucleic acids, certain guanine (G)-rich sequences have been shown to fold into non-canonical G-quadruplex structures formed through the stacking of Hoogsteen hydrogenbonded G-tetrads stabilized by a metal cation (Fig. 1a,b)1,2. The presence of DNA G-quadruplex structures in human cells, first demonstrated in ciliates3, is now firmly established4,5, which provides the basis for the elucidation of their function in normal and disease states6–8.
Figure 1.
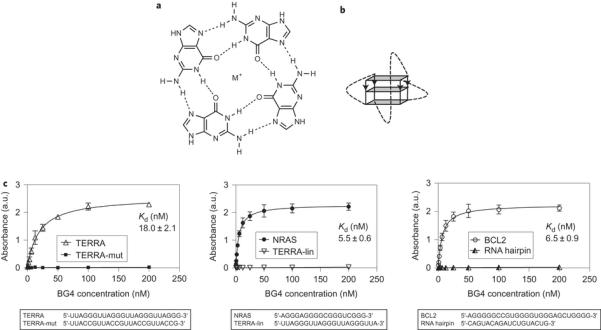
In contrast to DNA G-quadruplexes, little is known about RNA G-quadruplexes and there is a particular need to provide explicit evidence that G-quadruplex structures actually form in native cellular RNA. Biophysical studies support this possibility, as RNA G-quadruplexes form in vitro and are thermodynamically more stable than their DNA counterparts under near-physiological conditions9,10. The high-resolution structure of the telomeric repeatcontaining RNA (TERRA) G-quadruplex shows that the 2′-hydroxyl group imparts intramolecular hydrogen bonding within the G-quadruplex11 and contributes to the preference of RNA G-quadruplexes to adopt a parallel-stranded tertiary structure. This work also shed light on the potential for ligand-binding selectivity, and a crystal structure of TERRA bound to an acridine ligand revealed additional facets of RNA G-quadruplexes, such as loop involvement and hydrogen-bonding networks, not found in their DNA counterparts12. These unique chemical qualities of RNA G-quadruplexes suggest the possible development of smallmolecule ligands that are selective for RNA rather than for DNA G-quadruplexes13. Indeed, through a template-directed in situ ‘click chemistry’ approach, recently we discovered a small molecule, carboxypyridostatin (carboxyPDS), that exhibits high molecular specificity for RNA over DNA G-quadruplexes14.
Sequence-motif analyses have highlighted the enrichment of potential G-quadruplex-forming sequences within particular RNA domains important for nucleic acid function, including the first intron and 5′- and 3′-untranslated regions (UTRs)15,16. A number of biochemical and cell-based studies provided early insights into the possible consequences of RNA G-quadruplex formation in biology. For example, our work demonstrated that a G-quadruplex in the 5′-UTR of NRAS can inhibit translation17. More generally, 5′-UTR G-quadruplexes may be important regulatory elements that repress18 or promote19 translation. Inhibitory G-quadruplexes are also seen within coding sequences20, and G-quadruplexes located in internal ribosome entry sites have been shown to promote translation21,22. Several classes of G-quadruplex smallmolecule ligands, such as pyridostatins (PDS)23, cationic porphyrins24,25 and bisquinoliniums26,27, have been employed to modulate translation both in vitro and in cellular assays and are presumed to involve targeting of RNA G-quadruplexes. Also, it has been suggested that RNA G-quadruplexes mediate differential processing of transcripts, such as in IGF-II 3′-UTR endonucleolytic cleavage28, TP53 and BACE1 alternative splicing29,30, and LRP5 and FXR1 alternative polyadenylation31. They are also implicated in the transport and localization of certain RNA transcripts to different subcellular components, such as the plasma membrane and cytoskeleton32 or neurites33.
Naturally occurring proteins have been shown to exhibit RNA G-quadruplex-binding ability, which supports the endogenous formation of G-quadruplexes in cellular RNA. For example, the telomeric protein TRF2 binds to the TERRA G-quadruplex34, fragile X mental retardation protein (FMRP) binds to several transcripts containing a G-quadruplex motif, including its own RNA20,35,36, and Xenopus Pat1 proteins have been found to bind to RNA G-quadruplexes37. RHAU, a human DEAH-box helicase, also binds to RNA G-quadruplexes with high affinity and specificity38,39. To date, RHAU and DHX940 are the only helicases known to unfold RNA G-quadruplexes, which suggests that cells have endogenous mechanisms to resolve these highly stable structures.
Here we show that a G-quadruplex-specific antibody, BG4 (ref. 4), binds to RNA G-quadruplex structures and can be used to visualize RNA G-quadruplexes within the cytoplasm of human cells. We also demonstrate that G-quadruplex-specific small molecules stabilize endogenous G-quadruplex structures in cytoplasmic RNA within cells. Furthermore, we show that the RNA G-quadruplex-binding small molecule carboxyPDS increases the number of G-quadruplexes only in the cytoplasm, which suggests that in cells this ligand selectively traps cytoplasmic RNA G-quadruplexes and not nuclear DNA G-quadruplexes. Our findings provide substantive evidence for the presence of RNA G-quadruplex structures within the human transcriptome, and for their tractability as molecular targets for small-molecule ligands.
Results and discussion
Recognition of RNA G-quadruplex structures by a G-quadruplex-selective antibody. Previously, we developed G-quadruplex structure-specific antibodies4,41 by in vitro phage display methodologies. One such antibody, BG4, was employed to visualize DNA G-quadruplex structures in the nuclei of human cells4. Here, we investigated whether the BG4 antibody is also able to recognize RNA G-quadruplex structures, and whether it can be used to demonstrate robustly the presence of RNA G-quadruplexes in cells. An enzyme-linked immunosorbent assay (ELISA) approach was employed to show that BG4 does bind with low nanomolar affinity to several previously characterized RNA Gquadruplex structures, such as the TERRA34, BCL242 and NRAS17 G-quadruplexes (Kd: TERRA _18.0 nM, BCL2 _6.5 nM and NRAS _5.5 nM) (Fig. 1c). Using circular dichroism (CD) spectroscopy, we first confirmed that these oligonucleotides fold into stable G-quadruplex structures under the same conditions as used for the ELISA experiments Supplementary Fig. 1). We further confirmed BG4 G-quadruplex specificity by showing the absence of BG4 binding to a RNA hairpin or single-stranded RNA oligonucleotides (Fig. 1c). As single-stranded controls, we utilized two oligonucleotides derived from the TERRA G-quadruplex oligonucleotide: TERRA-mut, which has two mutated Gs in each repeat (sequence (UUACCG)4), and TERRA-lin, which lacks the three Gs within the fourth repeat (sequence (UUAGGG)3UUA) and therefore cannot fold into G-quadruplex structures34. As BG4 binding affinity for DNA and RNA G-quadruplexes is comparable4, this suggests that the antibody recognizes a common structural feature of G-quadruplexes. To date, no information is available about the precise molecular interactions that mediate BG4 binding to various G-quadruplexes. Ultimately, NMR spectroscopy or X-ray crystallography data of the antibody bound to the G-quadruplex may provide detailed structural insights.
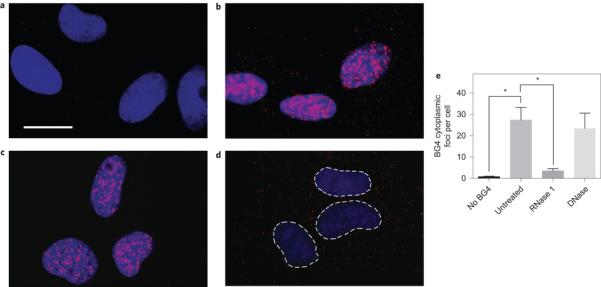
Visualization of RNA G-quadruplex structures in the cytoplasm of human cells. We next explored whether BG4 could be used to visualize RNA G-quadruplexes in human cells. To do this, we employed an optical microscopy approach that allowed us to fluorescently label and image endogenous G-quadruplex structures in a panel of normal (human umbilical vein endothelial cells (HUVEC)), immortalized (SV40-transformed MRC5 and GM847) and cancer (U2OS and HeLa) human cell lines (Fig. 2 and Supplementary Fig. 2). After fixation and permeabilization, cells were incubated with the BG4 antibody, and then an amplified fluorescence signal was generated by incubation with a secondary antibody that recognizes a Flag epitope tag present on BG4, followed by a tertiary fluorochrome-labelled antibody that binds to the secondary antibody, much as we previously described for the detection of DNA G-quadruplex structures4. Consistent with these earlier findings, we observed intense nuclear BG4 foci; however, on longer exposures, it was clear that less-intense punctate staining was also distributed throughout the cytoplasm of all cell lines examined (Fig. 2b and Supplementary Fig. 2). Although BG4 cytoplasmic foci were readily apparent by fluorescent microscopy, we further confirmed their cytoplasmic localization by staining with CellMask green, a fluorescent stain that reveals the cell boundaries (Supplementary Fig. 3). The presence of cytoplasmic foci was BG4 dependent, as no signal could be quantified (,1 cytoplasmic focus per cell) in the absence of the primary antibody (Fig. 2a,e). We then validated that BG4 cytoplasmic staining was indeed caused by RNA. When fixed permeabilized cells were treated with RNase A, prior to BG4 staining, we found that the foci in the cytoplasm disappeared (Fig. 2c,e). Importantly, this marked reduction was not observed on DNase treatment (Fig. 2d,e). In these experiments, each BG4 staining focus most probably represents the detection of a cluster of multiple G-quadruplexes within a single optically resolvable area. In our immunofluorescence experiments, usually cells were fixed with formaldehyde, a cross-linking fixative. However, to demonstrate that the method of fixation had no influence on the results, we also used an alternative fixation protocol with ethanol, a precipitating fixative, which gave comparable results (Supplementary Fig. 4). Although these results strongly support that the observed staining is not the result of a specific fixation artefact and that G-quadruplexes are structures intrinsic to endogenous RNA, we cannot rule out the possibility that the fixation process could influence the structure of the RNA. Taken together, our results show that the G-quadruplexes detected in the cytoplasm of human cells are present in endogenous RNA transcripts.
Figure 2.
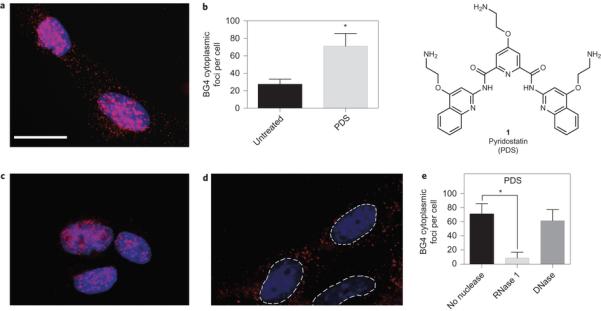
Small-molecule-mediated trapping of endogenous RNA G-quadruplexes in the cytoplasm of human cells. There is growing interest in the chemical biological intervention in biology and medicine through the targeting of G-quadruplex structures43,44. We therefore explored whether a small molecule could trap cytoplasmic RNA G-quadruplex structures in cells. The small molecule PDS (1) (Fig. 3), which binds both DNA and RNA G-quadruplexes in biophysical assays14,45, was used to treat cells for 24 hours prior to cell fixation and staining with BG4. Figure 3a,b shows that PDS treatment of cells causes a substantial increase (_2.4-fold) in the number of foci detected in the cytoplasm, which are sensitive to RNase but not to DNase treatment (Fig. 3c–e). Independently, we confirmed by ELISA measurements that BG4 binding affinity for DNA or RNA Gquadruplex structures is unchanged in the presence of the stabilizing ligand PDS4 (Supplementary Fig. 5). These results demonstrate that a small-molecule ligand can target and trap RNA G-quadruplex structures in the cytoplasm of human cells.
Figure 3.
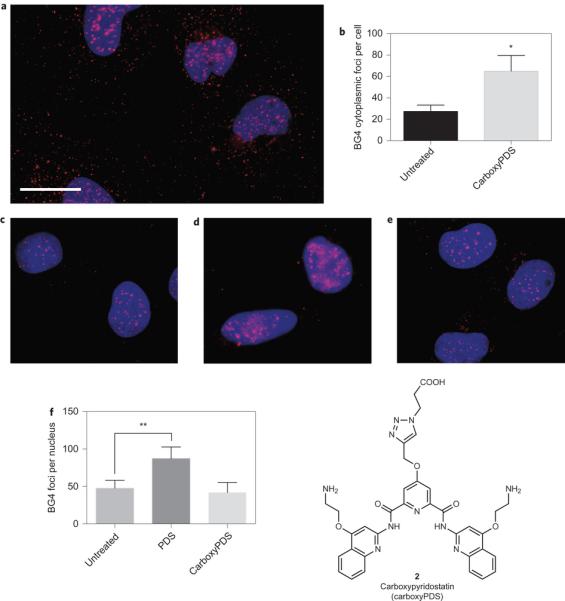
Application in cells of a RNA G-quadruplex-selective ligand reveals structural discrimination of endogenous G-quadruplexes. Recently, we identified by an in situ ‘click chemistry’ approach a small molecule, carboxyPDS (2), that shows a preference for RNA G-quadruplexes as compared to DNA G-quadruplexes14. This selectivity of carboxyPDS could thus be employed to substantiate the presence of RNA G-quadruplex structures in human cells. We therefore used BG4-based quantification of DNA (nuclear) and RNA (cytoplasmic) G-quadruplexes to evaluate whether carboxyPDS selectively traps RNA G-quadruplexes within a cellular context (Fig. 4). We first confirmed by ELISA measurements that BG4 binding affinity for RNA G-quadruplexes is unaltered by carboxyPDS (Supplementary Fig. 5). On treatment of human cells with carboxyPDS, we observed a marked increase (_2.3-fold) in the number of BG4 cytoplasmic foci (Fig. 4a,b), which is comparable to that following PDS treatment (Fig. 3a,b) and is indicative of the presence of a higher number of RNA G-quadruplexes. Strikingly, carboxyPDS did not cause an increase in the number of BG4 nuclear foci, and this is in contrast to PDS, which traps endogenous DNA G-quadruplex structures and leads to more BG4 staining in the nuclei (Fig. 4c–f )4. Similar to our previous observations4, we confirmed that BG4 targets in the nucleus are, indeed, DNA structures, as BG4 nuclear foci are sensitive to DNase but not to RNase treatment in PDS- or carboxyPDS-treated cells (Supplementary Fig. 6). The finding that carboxyPDS does not cause stabilization of nuclear DNA G-quadruplex structures is consistent with our earlier studies that show carboxyPDS is unable to trap telomeric G-quadruplexes and displace the telomeric protein TRF1 from telomeres, whereas PDS does both14. These results confirm that the RNA versus DNA selectivity of a G-quadruplex ligand, demonstrated biophysically in vitro, can be preserved within a cellular context.
Figure 4.
Conclusions
In summary, our findings provide substantive evidence for the formation of RNA G-quadruplexes in the cytoplasm of human cells. Given the studies and hypotheses implicating RNA G-quadruplexes as structural elements linked to RNA functions, our observations provide cell-based structural evidence that has so far been absent in the field. Our finding that RNA G-quadruplexes can be trapped in cells by G-quadruplex-specific ligands supports the prospect of intervention in biological processes via targeting such structures with chemical probes. The demonstration of preferential targeting of endogenous RNA G-quadruplexes is proof-of-concept that strategies based on small molecules have the potential to decouple the recognition of RNA G-quadruplexes from DNA G-quadruplexes in cells. The ability to visualize and quantitate DNA G-quadruplexes4, and now RNA G-quadruplexes, in human cells opens up new experimental strategies to explore the dynamics and functions of this fascinating class of nucleic acid structures in a biologically relevant context.
Methods
G-quadruplex structure-specific antibody. Isolation and production of the singlechain phage display antibody BG4 was performed as described in Biffi et al4. CD spectroscopy. For each experiment, 5 mM oligonucleotides (Table 1) were annealed in 10 mM Tris HCl, pH 7.4, 100 mM KCl by slow cooling from 95 8C to 4 8C. CD spectra were recorded on an Applied Photo-physics Chirascan CD spectropolarimeter using a 1 mm path-length quartz cuvette. CD measurements were performed at 25 8C over a range of 200–320 nm using a response time of 1 s, 1 nm step and 0.5 nm bandwidth. The recorded spectra represent a smoothed average of three scans, zero-corrected at 320 nm and normalized (molar ellipticity is quoted in 105 deg cm2 dmol21). The absorbance of the buffer was subtracted from the recorded spectra. In melting experiments, oligonucleotides were heated from 25 8C up to 95 8C at a rate of 1 8C min21, and spectra were recorded at intervals of 5 8C. Melting curves were obtained by plotting normalized molar ellipticity recorded at 265 nm using GraphPad Prism (GraphPad Software). ELISA experiments. These were carried out to measure the binding affinity and specificity and were performed using standard methods. Briefly, biotinylated oligonucleotides (Table 1) (biomers.net GmbH) were bound to streptavidin-coated plates followed by incubation with BG4, and detection was achieved with an anti-Flag horseradish peroxidase (HRP)-conjugated antibody (ab1238, Abcam) and 3,3′,5,5′-tetramethylbenzidine (HRP substrate, Roche). Signal intensity was measured at 450 nm on a PHERAstar microplate reader (BMG Labtech). Dissociation constants were calculated from binding curves using GraphPad Prism (GraphPad Software). Error bars represent standard error of mean calculated from three replicates. G-quadruplex oligonucleotides were folded in 10 mM Tris HCl, pH 7.4, 100 mM KCl by slow cooling from 95 8C to 4 8C. Cell cultures and immunofluorescence. U2OS (osteosarcoma), HeLa (cervical carcinoma), SV40-transformed GM847 (human skin fibroblasts) and SV40-transformed MRC-5 (fetal lung fibroblasts) cells were cultured in DMEM (Gibco), 1% l-glutamine, 10% fetal bovine serum; HUVEC cells (human umbilical vein endothelial cells) were cultured in endothelial cell growth medium (Lonza) at 37 8C with 5% CO2. Cells grown on glass coverslips were fixed in 2% paraformaldehyde/PBS or in ethanol (Supplementary Fig. 4) and permeabilized with 0.1% Triton X-100/PBS. After blocking in 2% MarvelTM/PBS, immunofluorescence was performed using standard methods, incubating at 37 8C with BG4, followed by anti-Flag (No. 2368, Cell Signaling Technology) and anti-rabbit Alexa 594-conjugated (A11037, Life Technologies) antibodies. Coverslips were mounted with Prolong Gold/4′,6-diamidino-2-phenylindole (DAPI; Life Technologies). Cell boundaries were highlighted using CellMask green stain (Life Technologies) (Supplementary Fig. 3). For enzymatic treatments, coverslips were incubated after permeabilization with 0.12 U ml21 Turbo DNase (Ambion) or with 100 mg ml21 RNase A (Fermentas) for one hour at 37 8C. For G-quadruplex ligand treatments, PDS was synthesized as described in Muller et al.46 and carboxyPDS as in Di Antonio et al.14. Cells were incubated with 2 mM of compound for 24 hours before fixation. Digital images were recorded with a DP70 camera (Olympus) on an Axioskop 2 plus microscope (Zeiss). For cytoplasmic foci detection, exposure was increased relative to that used for detection of nuclear foci and kept constant between different replicates. Foci number was quantified by image analysis using Volocity software (Perkin Elmer) with 100–200 cells counted per condition and the standard error of the mean calculated from three replicates. Statistical analyses and P values were calculated using the Student’s t-test. Frequency-distribution graphs were plotted using GraphPad Prism (GraphPad Software).
Table 1. Oligonucleotides used in the CD and ELSIA experiments.
| Oligonucleotide | Sequence |
|---|---|
| RNA hairpin | 5′CAGUACAGAUCUGUACUG-3′ |
| TERRA34 | 5′-(UUAGGG)4-3′ |
| TERRA-mut34 | 5′-(UUACCG)4-3′ |
| TERRA-lin34 | 5′-(UUAGGG)3UUA-3′ |
| NRAS17 | 5′-AGGGAGGGGCGGGUCUGGG-3′ |
| BCL242 | 5′-AGGGGGCCGUGGGGUGGGAGCUGGGG-3′ |
Supplementary Material
Supplementary information
Acknowledgements
We thank D. Sanders for stimulating discussions and Cancer Research UK for funding.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guaninerich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 2.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guaninequadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nature Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nature Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nature Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nature Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacca B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang AY, Bugaut A, Balasubramanian S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry. 2011;50:7251–7258. doi: 10.1021/bi200805j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collie GW, Haider SM, Neidle S, Parkinson GN. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collie GW, Sparapani S, Parkinson GN, Neidle S. Structural basis of telomeric RNA quadruplex – acridine ligand recognition. J. Am. Chem. Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- 13.Collie G, et al. Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem. Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 14.Di Antonio M, et al. Selective RNA versus DNA G-quadruplex targeting by in situ click chemistry. Angew. Chem. Int. Ed. 2012;51:11073–11078. doi: 10.1002/anie.201206281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nature Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwala P, Pandey S, Mapa K, Maiti S. The G-quadruplex augments translation in the 5′ untranslated region of transforming growth factor b2. Biochemistry. 2013;52:1528–1538. doi: 10.1021/bi301365g. [DOI] [PubMed] [Google Scholar]
- 20.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 22.Bonnal S, et al. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugaut A, Rodriguez R, Kumari S, Hsu ST, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010;8:2771–2776. doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris MJ, Wingate KL, Silwal J, Leeper TC, Basu S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012;40:4137–4145. doi: 10.1093/nar/gkr1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faudale M, Cogoi S, Xodo LE. Photoactivated cationic alkyl-substituted porphyrin binding to g4-RNA in the 5′-UTR of KRAS oncogene represses translation. Chem. Commun. 2012;48:874–876. doi: 10.1039/c1cc15850c. [DOI] [PubMed] [Google Scholar]
- 26.Gomez D, et al. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halder K, Largy E, Benzler M, Teulade-Fichou MP, Hartig JS. Efficient suppression of gene expression by targeting 5′-UTR-based RNA quadruplexes with bisquinolinium compounds. Chem Bio Chem. 2011;12:1663–1668. doi: 10.1002/cbic.201100228. [DOI] [PubMed] [Google Scholar]
- 28.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisette JF, Montagna DR, Mihailescu MR, Wolfe MS. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J. Neurochem. 2012;121:763–773. doi: 10.1111/j.1471-4159.2012.07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcel V, et al. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 31.Beaudoin JD, Perreault JP. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013;41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd AG, Lin H, Ebert AD, Liu Y, Androphy EJ. COPI transport complexes bind to specific RNAs in neuronal cells. Hum. Mol. Genet. 2013;22:729–736. doi: 10.1093/hmg/dds480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian M, et al. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011;12:697–704. doi: 10.1038/embor.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biffi G, Tannahill D, Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 36.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 37.Marnef A, et al. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA. 2010;16:2094–2107. doi: 10.1261/rna.2295410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creacy SD, et al. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaughn JP, et al. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair. 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Fernando H, Rodriguez R, Balasubramanian S. Selective recognition of a DNA G-quadruplex by an engineered antibody. Biochemistry. 2008;47:9365–9371. doi: 10.1021/bi800983u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahid R, Bugaut A, Balasubramanian S. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry. 2010;49:8300–8306. doi: 10.1021/bi100957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Antonio M, Rodriguez R, Balasubramanian S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods. 2012;57:84–92. doi: 10.1016/j.ymeth.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collie GW, Parkinson GN. The application of DNA and RNA Gquadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez R, et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller S, et al. Pyridostatin analogues promote telomere dysfunction and longterm growth inhibition in human cancer cells. Org. Biomol. Chem. 2012;10:6537–6546. doi: 10.1039/c2ob25830g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information