PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane (original) (raw)
PDZ domains have recently emerged as central organizers of protein complexes at the plasma membrane. Although new information about PDZ domains appears at a bewildering rate, we will attempt to distill a few general concepts that have emerged about their biology. These include the structural basis for specificity of their binding interactions and ideas about how they organize both small local protein complexes used for signal transduction (transducisomes) and larger two-dimensional complexes like cell junctions and plasma membrane domains.
Polarized epithelial cells have discrete apical, basal-lateral, and junctional membrane domains. Each domain has a specific molecular composition, which includes protein complexes composed of distinct transmembrane, membrane-associated, and cytosolic components. These protein complexes mediate the adhesive properties of particular cells, the formation of the paracellular barrier (tight junctions), ion transport, and transmission of signals between adjacent cells that regulate growth, differentiation, and homeostasis.
Formation of these protein complexes is determined in large part by the interactions of modular protein-binding domains. These are structurally conserved elements with unique molecular specificities that can be found in proteins of many different functions. Examples of these domains include SH3 domains, which recognize amino acid sequence variations around a basic Pro-X-X-Pro site; the SH2 and PTB domains, which recognize phosphotyrosine and contiguous residues; and PDZ domains (reviewed in ref. 1). Because binding specificities are based on a few amino acid residues, these domains are uniquely suited to permit evolution of new protein interactions by coordinate mutations in the domain and target peptide sequence. These domains are, figuratively speaking, the glue that binds protein complexes together, and their unique specificity and regulated binding determine the distinct compositions of different complexes.
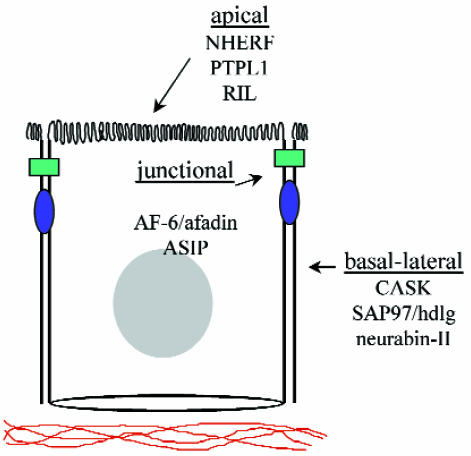
PDZ domains were originally identified as conserved sequence elements within the postsynaptic density protein PSD95/SAP90, the Drosophila tumor suppressor dlg-A, and the tight junction protein ZO-1. Although originally referred to as GLGF or DHR motifs, they are now known by an acronym representing these first three PDZ-containing proteins (PDZ: PSD95/DLG/ZO-1). These 80–90 amino acids sequences have now been identified in well over 75 proteins and are characteristically expressed in multiple copies within a single protein. They are also found throughout phylogeny in organisms as diverse as metazoans, plants, and bacteria (2). Such a broad species distribution appears to be unique to this domain, but perhaps the most distinguishing feature of PDZ domains is the observation that the overwhelming majority of proteins containing them are associated with the plasma membrane. Although PDZ domains are found in many different structures, each PDZ protein is generally restricted to specific subcellular domains, such as synapses; cell–cell contacts; or the apical, basal, or lateral cell surface (Fig. 1). This leads to the speculation that PDZ domains evolved early to provide a central role in the organization of plasma membrane domains.
Figure 1.
Schematic diagram of a vertebrate polarized epithelial cell, showing the distinct subcellular localization of several epithelial PDZ proteins into the apical, basal-lateral, and junctional domains.
Mechanism of protein binding and the basis for specificity.
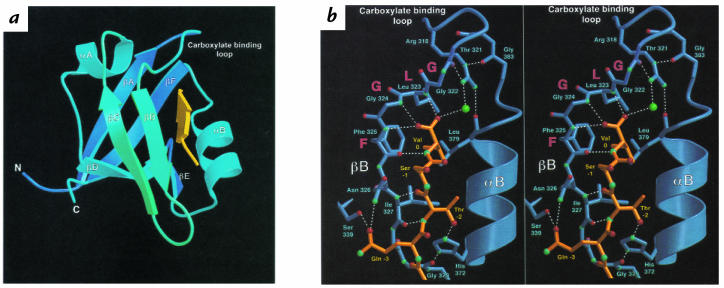
The original demonstration that PDZ domains were indeed protein-binding motifs came from the observation that the COOH-terminal tail of the Shaker K+ channel could bind to the second PDZ domain of the synaptic protein PSD95 both in vitro and in vivo (3). Mutational analysis demonstrated that this interaction required the COOH-terminal sequence (T/S)XV, where serine or threonine are permitted at the –2 position, any residue could substitute at the –1, and valine or a hydrophobic side chain must be present at the 0 (or COOH-terminal) position. The basis for this specificity became apparent when the crystallographic structure of this domain was solved in a complex with its peptide ligand (4). This structure demonstrates that the PDZ domain is composed of six β strands (βA–βF) forming two opposing antiparallel sheets flanked by two α helices (αA and αB) (Fig. 2a). The peptide, or COOH-terminus of the interacting protein, lies within an elongated surface groove as an antiparallel β strand interacting with the βB strand and the αB helix. The hydrophobic COOH-terminal side chain inserts into a hydrophobic cavity at the end of the groove, where the carboxylate group of the terminal valine is coordinated by hydrogen bonding to molecules of the carboxylate-binding loop. Interactions between the peptide and the βB strand involve only main chain bonds, suggesting a general, sequence-independent set of interactions that may be common to all PDZ-target pairs (Fig. 2b). Specificity is determined on the opposite side of the groove by interaction of side chain residues at the –2 position (S/T hydroxyl) with the positively charged histidine side chain at the base of helix αB (Fig. 2b). Several other binding pairs have now been defined in which the –2 position side chain is hydrophobic and interacts with a corresponding hydrophobic residue at the base of αB. PDZ domains recognizing S/T or a hydrophobic residue at the –2 position have been termed class I and class II, respectively.
Figure 2.
Structural diagrams of the PDZ3 domain of PSD95, depicting interactions with a peptide ligand. (a) Ribbon diagram of the PDZ3 domain. The peptide (yellow) lies in the groove created by the βB strand and the αB helix, and the COOH-terminus of the peptide lies in the hydrophobic activity created by the βA–βB or carboxylate loop. (b) Chemical interactions involved in peptide binding. Stereoscopic images depicting hydrogen bonding (dashed white lines) between residues of the PDZ domains (blue) and the peptide ligand (orange). Oxygen atoms are shown in red and nitrogen atoms in blue. The green sphere is a well-ordered water molecule linking the carboxylate group to Arg318. Reproduced, with permission, from ref. 4.
This structural model (Fig. 2) would predict that specificity is based primarily on interactions mediated by only two residues within the COOH-terminal ligand, which seems an improbable mechanism for generating binding specificity between a large and growing number of binding partners. This dilemma may be resolved in two ways. First, PDZ domains are always found within larger multidomain proteins. Other domains or sequences may target the protein to a specific membrane domain, such as the lateral membrane surface, thereby limiting the number of potential targets the PDZ may encounter. In some cases, these targeting sequences bind to actin-binding proteins, such as protein 4.1, that already have restricted spatial distributions. Second, the determination of binding specificity is likely to depend on more than just residues at the –2 and 0 positions. Analysis of target specificity using degenerate peptide and phage display library screening methods has demonstrated that many PDZ interactions require additional side chain interactions at the –1, –4, and/or –5 positions (5, 6). Many of the specificities defined by peptide library screening and confirmed by in vivo binding assays are difficult to explain on the basis our present understanding of crystallographic data on class I and II domains. Thus, a further understanding of PDZ domain specificity will require continued definition of novel binding interactions along with crystallographic or nuclear magnetic resonance studies.
Adding to the complexity of how PDZ domains create protein networks is the recognition that some bind to non–COOH-terminal target sequences. For example, PDZ domains have also been demonstrated to interact with internal (T/S)XV motifs, such as that described in the Drosophila phototransduction system between the PDZ protein InaD and the transient receptor potential (TRP) store-operated Ca2+ channel (7). PDZ domains have also been demonstrated to bind directly to other PDZ domains, forming homomeric and heteromeric complexes (8). Finally, PDZ interactions have been documented with recognized domains commonly found in other proteins of the cortical cytoskeleton, such as LIM domains or ankyrin and spectrin repeats. The structural basis for these interactions is unknown. It is possible that the PDZ domain is binding to a short polypeptide with standard consensus embedded within the larger motif, similar to the interaction between InaD and TRP. Alternatively, these target sequences may interact with a unique surface on the PDZ domain. The latter hypothesis is supported by the observation that some PDZ domains can dimerize while simultaneously binding to the COOH-terminal motif of a third protein and that deletions within the PDZ domain that eliminate COOH-terminal binding can have no effect on PDZ dimerization (9). It is unclear whether all PDZ domains share multiple protein-binding mechanisms or whether these examples represent interactions unique to specific PDZ domains. In any case, they point to the extremely complex way in which PDZ domains are used to connect transmembrane proteins with the cortical protein network.
Recent observations suggest that the interaction of PDZ domains with transmembrane binding partners can also be directly regulated by posttranslational modification or by receptor activation. For example, binding of the inwardly rectifying K+ channel Kir 2.3 to the second PDZ domain of PSD-95 is inhibited by protein kinase A phosphorylation of the serine residue at the –2 position in the COOH-terminus of Kir 2.3 (10). From the structural model, it is easy to see how addition of the large charged phosphate group to the side chain at position –2 would interrupt binding to the cognate histidine residue within the αB helix of the PDZ domain. Other regulated interactions are presently more difficult to explain and imply some target proteins undergo conformational changes before they can engage the PDZ domain. For example, the interaction of the β2-adrenergic receptor with the epithelial PDZ protein NHERF is dependent on agonist-mediated activation of the receptor (11), and interaction of the EphB3 receptor tyrosine kinase with the PDZ protein AF-6 is dependent on the kinase activity of the receptor (6). It seems reasonable to predict that regulated association with PDZ-containing proteins will turn out to be a general mechanism for regulating the temporal and spatial assembly of PDZ-containing complexes, the activity of transmembrane proteins, and the propagation of signal transduction events across and along the plasma membrane.
PDZ domains mediate the formation of protein complexes at the plasma membrane.
The most general function of PDZ domains may be to localize their ligands to the appropriate plasma membrane domain. In polarized epithelial cells, PDZ proteins clearly localize at distinct apical, basal-lateral, and junctional membrane domains (Fig. 1) and, in most cases, colocalize with their transmembrane and cytosolic binding partners. For example, the PDZ protein NHERF both binds to and colocalizes with the cystic fibrosis transmembrane regulator (CFTR) chloride channel and the Na+/H+ exchanger on the apical plasma membrane (Fig. 3b and ref. 12). Similarly, in Drosophila, the DLG protein colocalizes with one of its subcellular ligands, fasciclin-III, at septate junctions of imaginal disk epithelia and is required for the proper localization of this protein, aswell as that of the protein 4.1 homolog coracle, to septate junctions (13). In Caenorhabditis elegans, a complex of three PDZ proteins — lin2, lin7, and lin10 — is required for the appropriate localization of the receptor tyrosine kinase let23 to the basal-lateral surface of the vulval precursor cells. The PDZ domain in one of these proteins, lin7, binds directly to the COOH-terminal tail of let23. Mutations in lin7, as well as in lin2 or lin10, result in mislocalization of let23 to the apical plasma membrane (14).
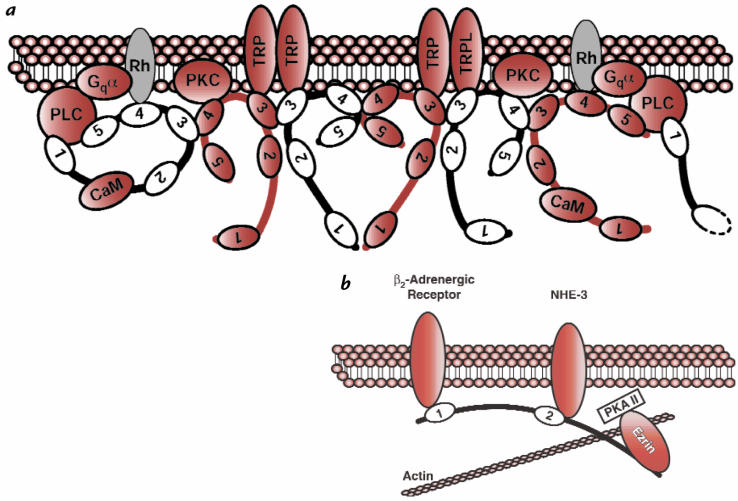
Figure 3.
Schematic diagram of InaD and NHERF, illustrating the protein–protein interactions mediated by these two PDZ proteins. (a) InaD binds to the G protein (Gqα)–linked receptor rhodopsin (Rh), the light-activated cation channels TRP and TRPL, phospholipase C-β (PLC), and protein kinase C (PKC) via PDZ domains. Calmodulin binds to the region between PDZ1 and PDZ2. A higher-order structure is organized at the plasma membrane by homomeric and heteromeric binding between PDZ domains 3 and 4. Reproduced, with permission, from ref. 9. (b) The β2-adrenergic receptor binds to the first PDZ domain of NHERF, and the Na+/H+ exchanger NHE-3 binds to a region adjacent to and including the second PDZ domain. Ezrin binds to a domain in the COOH-terminus of NHERF, linking the protein complex to the actin cytoskeleton and protein kinase A (PKA-II). Both PKA-II and the β2-adrenergic receptor regulate the activity of NHE-3.
PDZ proteins also clearly have a fundamental role spatially clustering and anchoring transmembrane proteins within specific subcellular domains. This organizational capacity was first demonstrated by expression studies of PSD-95 and one of its transmembrane binding partners, the Shaker K+ channel. Kim et al. (3) demonstrated that expression of both proteins in a heterologous cell type resulted in coclustering of the PDZ protein and its ligand on the cell surface and that this clustering was eliminated when the PDZ-binding motif in the K+ channel was mutated (3). This property has now been demonstrated in several PDZ proteins and their transmembrane ligands, both neuronal and epithelial. The in vivo significance of PDZ-mediated interactions in the organization of transmembrane proteins is aptly demonstrated by the Drosophila DLG protein, a homolog of PSD-95 that is present in both neuronal and epithelial cells. DLG can also crosslink the Shaker K+ channel in a heterologous cell assay, and mutation of either DLG or the PDZ-binding domain of the Shaker K+ channel disrupts channel clustering at the synapse (15).
The ability to cluster transmembrane receptors and channels has great functional significance. Receptor clustering is a well-established prerequisite for receptor activation and might also provide a mechanism to coordinate activity of different transmembrane receptors and ion channels. An example of this is provided by NHERF, an epithelial PDZ protein localized to the apical plasma membrane that was originally identified as a cofactor required for protein kinase A–mediated regulation of the Na+/H+ exchanger NHE-3 (16). NHERF has two PDZ domains and binds directly to both the β2-adrenergic receptor and NHE-3. Investigators have demonstrated that agonist-mediated activation of β2-adrenergic receptor can alter the activity of NHE and that NHERF is required for this regulation (11). Thus, PDZ-mediated clustering may activate and coordinate the activity of different plasma membrane proteins (Fig. 3).
PDZ interactions are not limited to transmembrane proteins but also include a large number of different cytosolic proteins that are recruited to membrane complexes through PDZ-mediated interactions. Perhaps the best example of this property is provided by the syntrophins and neuronal nitric oxide synthase (nNOS), both of which contain PDZ domains. Syntrophins are membrane-associated components of the dystrophin-associated protein complex in muscle cells that bind to nNOS via interaction between the PDZ domains in these proteins. Significantly, nNOS isoforms lacking a PDZ domain fail to localize to the plasma membrane (8), and nNOS is diffusely distributed throughout the cytosol in mice lacking the syntrophin gene (17). Thus, PDZ domains, like those found in syntrophin and nNOS, have the capacity to serve as adapters or linkers between transmembrane and cytosolic proteins, recruiting cytosolic proteins to specific subcellular domains.
The transducisome: organization of the components of cellular signaling pathways.
Many of the cytosolic proteins that interact with membrane-associated PDZ proteins are components of signal transduction pathways. These include protein kinases, regulators of G proteins, phospholipase C, and GTPase-activatin g proteins for small GTP-binding proteins such as Rac and Rho. This suggests that PDZ domains can serve as a conduit for coupling channels and transmembrane receptors to downstream signaling elements, and this is nowhere better illustrated than in the PDZ protein InaD (Fig. 3a). InaD is a component of the Rhabdomere of Drosophila photoreceptor cells and contains five PDZ domains. Through a series of genetic and biochemical experiments, investigators have determined that most of the proteins involved in the G protein–mediated signal cascade required for phototransduction bind to one or more of the PDZ domains in InaD (reviewed in ref. 18). Thus, InaD not only interacts with the transmembrane proteins rhodopsin and the TRP Ca2+ channel but also with protein kinase C and phospholipase C-β via one or more of these five PDZ domains. These interactions are required for both localization of these proteins to the plasma membrane (Rhabdomere) and effective phototransduction. InaD organizes these proteins into a highly organized signaling complex, appropriately termed the transducisome (19). Organization of transducisomes by multidomain PDZ proteins will probably emerge as a common feature of cell organization.
Several aspects of this molecular organization are of great functional significance. First, the efficiency of signal transmission is increased by physically coupling receptors and downstream signaling molecules (or substrates). Second, the fidelity of the response to a specific signal is maintained. Because many of the downstream signaling molecules are used in other cellular pathways, physically coupling them to their receptors may isolate the response from crossover into these other pathways. As mentioned above, physical clustering of different receptors and ion channels could also allow for coordination of their respective activities. In the case of InaD, the experimental evidence suggests that spatial coupling of these two activities is required for wild-type phototransduction (7). Calcium influx through the TRP Ca2+ channel is required for feedback regulation of phototransduction through rhodopsin; thus, proximity of the TRP Ca2+ channel to rhodopsin may have functional consequences. Finally, the recent identification of the Ephrin receptor tyrosine kinases as ligands for several PDZ proteins raises the possibility that PDZ proteins may also recruit substrates to their membrane receptor kinases (20). Thus, by acting as scaffolds for cellular signaling proteins, PDZ domains may physically organize signal transduction pathways at the plasma membrane.
Mechanism of complex formation at the plasma membrane.
How do PDZ proteins contribute to the organization of complex cellular structures like synapses and the epithelial junctional complexes, which often contain several different PDZ proteins and their cellular ligands? Because a single PDZ protein contains anywhere from 1 to 13 PDZ domains, it is possible that a single PDZ protein could bind several copies of a particular transmembrane protein in a linear array, thereby clustering them into a single complex. However, the individual PDZ domains within a protein often show distinct specificities, such that only one receptor of a specific type can bind to any single PDZ protein. In addition, PDZ proteins with single PDZ domains are also effective at clustering transmembrane proteins in heterologous cell assays (20). Furthermore, while the InaD protein has the potential to bind all of the components of the G protein–mediated signal cascade, one InaD molecule does not have enough PDZ domains to bind them all at the same time, especially since several of these proteins bind to the same PDZ domain. Thus, while the presence of multiple PDZ domains within a single polypeptide may partly explain the ability to organize several distinct proteins into a single structure, it does not adequately explain the organization of these proteins into more complex cellular structures.
What then accounts for this higher order of complexity? At least one explanation may be provided by the ability of many PDZ proteins to form higher-order homomeric and heteromeric complexes. In some cases, this appears to be mediated by the PDZ domains themselves. As described above, PDZ domains can mediate direct interaction with other PDZ domains. The ability of the third and fourth PDZ domains in InaD to form both heteromeric and homomeric interactions has been proposed as a mechanism by which the InaD protein forms a higher-order scaffold for the components of the phototransduction pathway (Fig. 3a and ref. 18). Other PDZ proteins have been demonstrated to form cysteine bonds through conserved cysteine residues, and these bonds have been demonstrated to mediate the formation of multimeric complexes at the plasma membrane (21). Finally, other binding partners for several PDZ proteins have been identified that promote clustering, although the mechanism is currently unknown (22).
Another factor that is likely to affect the formation of higher-order complexes is the ability of many PDZ proteins to interact with the cortical cytoskeleton. Some PDZ proteins, such as the tight junction proteins ZO-1 and AF-6/afadin, appear to bind directly to actin filaments (23, 24). Others have been demonstrated to interact with members of the ERM family of actin-binding proteins. SAP97/hdlg had been demonstrated to bind to protein 4.1 through a unique domain known as the hook domain, whereas NHERF interacts with another ERM family member Ezrin (25, 26) through a novel motif in its COOH-terminus (Fig. 3b). More recently, a novel factor, CRIPT, has been identified that binds to the third PDZ domain of PSD95, which can mediate interactions with microtubules (27). These interactions are proposed to anchor these PDZ proteins to the cortical cytoskeleton, bringing them into close contact with other plasma membrane–associated proteins and adding stability to these protein microdomains.
Thus, higher-order networks can be mediated by multimerization of PDZ proteins and interaction with cytoskeleton. Other proteins can be recruited into this scaffold via direct or indirect interactions with other domains within the PDZ protein. In such a way, NHERF recruits Ezrin, which not only provides a link to the actin cytoskeleton but also binds to protein kinase A (Fig. 3 and ref. 28), which is required for the cAMP-mediated regulation of the Na+/H+ exchanger NHE-3 (29). All of these interactions may be dynamically regulated, either by posttranslation modifications, such as the interaction between PSD-95 and Kir 2.3 described above, or by competitive interactions between different PDZ-binding proteins for a single PDZ domain.
Future directions.
Recent experimental evidence using the genetics of Drosophila, C. elegans, and mice indicates that PDZ proteins are involved in the regulation of epithelial cell growth, differentiation, and morphogenetic movements during development. These systems will undoubtedly continue to provide great insight into the role PDZ proteins play in these phenomena. However, the precise nature of the molecular complexes mediated by PDZ proteins in epithelial tissues is still unresolved, and this remains an area of active investigation. Given the large number of PDZ-containing proteins and wide range of possible binding specificities, it seems likely many transmembrane proteins, ion channels, and receptors will be organized and regulated by PDZ domain complexes.
Acknowledgments
The authors would like to thank Christina Van Itallie for a critical reading of the manuscript. The authors are supported by grants DK-38979, DK-45134, and CA-66263 from the National Institutes of Health.
References
- 1.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Phillips C, Davies KE, Blake DJ. PDZ domains: targeting signaling molecules to sub-membranous sites. Bioessays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 3.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng S. Clustering of the Shaker-type K+ channels by direct interaction with the PSD-95/SAP90 family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 4.Doyle DA, et al. Crystal structure of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 5.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 6.Hock B, et al. PDZ-domain-mediated interaction of the Eph-related receptor tyrosine kinase EphB3 and the ras-binding protein AF6 depends on the kinase activity of the receptor. Proc Natl Acad Sci USA. 1998;95:9779–9784. doi: 10.1073/pnas.95.17.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shieh BH, Zhu MY. Regulation of the TRP channel by INAD in Drosophila photoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 8.Brenman JE, et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and a1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 9.Xu XZ, Choudhury A, Li X, Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen NA, Brenman JE, Snyder SH, Bredt DS. Binding of the inward rectifier K+ channel Kir 2.3 to PSD-95 is regulated by protein kinase a phosphorylation. Neuron. 1996;17:759–767. doi: 10.1016/s0896-6273(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 11.Hall R, et al. The B2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature. 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 12.Short D, et al. An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 13.Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaech S, Whitfield C, Kim S. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tejedor FJ, et al. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinman EJ, Steplock D, Wang Y, Shenolikar S. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na+-H+ exchanger. J Clin Invest. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kameya S, et al. Alpha 1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration. J Biol Chem. 1999;274:2193–2200. doi: 10.1074/jbc.274.4.2193. [DOI] [PubMed] [Google Scholar]
- 18.Montell C. TRP trapped in fly signaling web. Curr Opin Neurobiol. 1998;8:389–397. doi: 10.1016/s0959-4388(98)80066-4. [DOI] [PubMed] [Google Scholar]
- 19.Tsunoda S, et al. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 20.Torres RJ, et al. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their Ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 21.Hsueh YP, Kim E, Sheng M. Disulfide-linked head-to-head multimerization in the mechanism of ion channel clustering by PSD-95. Neuron. 1997;18:803–814. doi: 10.1016/s0896-6273(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 22.Kim E, et al. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanning AS, Jameson B, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. 1998;273:29745–53. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 24.Mandai K, et al. Afadin: a novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J Cell Biol. 1997;139:517–528. doi: 10.1083/jcb.139.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lue RA, Brandin E, Chan EP, Branton D. Two independent domains of hDlg are sufficient for subcellular targeting: The PDZ1-2 conformational unit and an alternatively spliced domain. J Cell Biol. 1996;135:1125–1137. doi: 10.1083/jcb.135.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reczek D, Berryman M, Bretscher A. Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the Ezrin-Radixin-Moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niethammer M, et al. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 28.Dransfield DT, et al. Ezrin is a cyclic AMP-dependent protein kinase anchoring protein. EMBO J. 1997;16:35–43. doi: 10.1093/emboj/16.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamprecht G, Weinman EJ, Yun C-HC. The role of NHERF and E3KARP in the cAMP-mediated inhibition of NHE3. J Biol Chem. 1998;273:29972–29978. doi: 10.1074/jbc.273.45.29972. [DOI] [PubMed] [Google Scholar]