Differential roles of microglia and monocytes in the inflamed central nervous system (original) (raw)
Phagocytic monocyte-derived macrophages associate with the nodes of Ranvier and initiate demyelination while microglia clear debris and display a suppressed metabolic gene signature in EAE.
Abstract
In the human disorder multiple sclerosis (MS) and in the model experimental autoimmune encephalomyelitis (EAE), macrophages predominate in demyelinated areas and their numbers correlate to tissue damage. Macrophages may be derived from infiltrating monocytes or resident microglia, yet are indistinguishable by light microscopy and surface phenotype. It is axiomatic that T cell–mediated macrophage activation is critical for inflammatory demyelination in EAE, yet the precise details by which tissue injury takes place remain poorly understood. In the present study, we addressed the cellular basis of autoimmune demyelination by discriminating microglial versus monocyte origins of effector macrophages. Using serial block-face scanning electron microscopy (SBF-SEM), we show that monocyte-derived macrophages associate with nodes of Ranvier and initiate demyelination, whereas microglia appear to clear debris. Gene expression profiles confirm that monocyte-derived macrophages are highly phagocytic and inflammatory, whereas those arising from microglia demonstrate an unexpected signature of globally suppressed cellular metabolism at disease onset. Distinguishing tissue-resident macrophages from infiltrating monocytes will point toward new strategies to treat disease and promote repair in diverse inflammatory pathologies in varied organs.
Blood-derived monocytes and resident microglia can both give rise to macrophages in the central nervous system (CNS). In tissue sections, macrophages derived from these two distinct precursors are indistinguishable at the light microscopic level both morphologically and by surface markers. Using flow cytometry, microglia- and monocyte-derived macrophages can be isolated separately from CNS tissue lysates and expression profiling suggests distinct functional capacities (Gautier et al., 2012; Chiu et al., 2013; Butovsky et al., 2014).
Microglia and monocytes are ontogenetically distinct: microglia derive from yolk-sac progenitors during embryogenesis (Ginhoux et al., 2010; Schulz et al., 2012), whereas monocytes continuously differentiate throughout postnatal life from bone marrow hematopoietic stem cells (HSCs), which require the transcription factor Myb. Microglial precursors are Myb independent, and microglia self-renew independently of bone marrow HSCs (Gomez Perdiguero et al., 2013). Distinct developmental origin and renewal mechanisms imply that monocyte-derived macrophages (MDMs) and microglia-derived macrophages (MiDMs) might exert different functions in pathological processes. Microglia represent one instance of tissue-resident macrophages, which reside in all organs. Studying the CNS as compared with other organs may carry advantages for distinguishing tissue-resident myeloid cells from infiltrating monocytes during disease, as there is virtually no background trafficking of monocytes in the CNS parenchyma of healthy animals.
In EAE, which models inflammatory aspects of MS (Williams et al., 1994; Ransohoff, 2012), macrophages dominate the inflammatory infiltrates and their numbers correlate to EAE severity (Huitinga et al., 1990, 1993; Ajami et al., 2011). However the cellular mechanisms by which macrophages promote disease progression are uncertain. Whether MiDMs or MDMs are functionally distinct and whether the two cell types differentially initiate demyelination or promote repair (Steinman et al., 2002) also remains elusive (Bauer et al., 1995). In MS autopsy tissues, prominent macrophage accumulation correlates with active demyelination (Ferguson et al., 1997; Trapp et al., 1998). Based on kinetics of cell accumulation and differential marker expression, it’s estimated that 30–50% of activated macrophages in active MS lesions derive from microglia (Brück et al., 1995; Trebst et al., 2001). Therefore, differential functions of MDMs and MiDMs are relevant for human demyelinating disease.
To date, no research techniques have permitted distinction between monocytes and microglia in CNS tissue without irradiation chimerism or parabiosis, techniques that confound interpretation or impose practical limitations (Ajami et al., 2007, 2011; Ransohoff, 2007). When F4/80+ macrophages were isolated from CNS and analyzed by flow cytometry using cells from double-heterozygous _Ccr2_rfp::_Cx3cr1_gfp mice with EAE, GFP was expressed by CD45dim/Ly6C− microglia, whereas RFP was restricted to CD45high/Ly6C+ monocytes (Saederup et al., 2010; Mizutani et al., 2012). These findings suggested an approach to clarifying distinct roles of MDMs and MiDMs in EAE based on differential expression of GFP and RFP reporters. Here, we use that strategy to extend previous findings and address the hypothesis that MDMs and MiDMs exert different functions in neuroinflammation. We detected detailed ultrastructural characterization of MDMs and MiDMs at EAE onset.
Unexpectedly, this approach provided insight into the cellular basis for autoimmune demyelination, which has remained obscure despite >80 yr of study in the EAE model. Here we provide evidence that MDMs initiate demyelination, often at nodes of Ranvier. In contrast, phagocytic microglia appear relatively inert at disease onset. Results from expression profiling provided insight into mechanisms and signaling pathways underlying the disparate effector properties of MDMs and MiDMs in EAE. The distinct functions of tissue-resident myeloid cells as compared with infiltrating macrophages broadly underlie disease pathogenesis in manifold circumstances and also hold promise for innovative treatment strategies.
RESULTS
In the CNS of mice of EAE, MDMs and MiDMs exhibit different accumulation kinetics
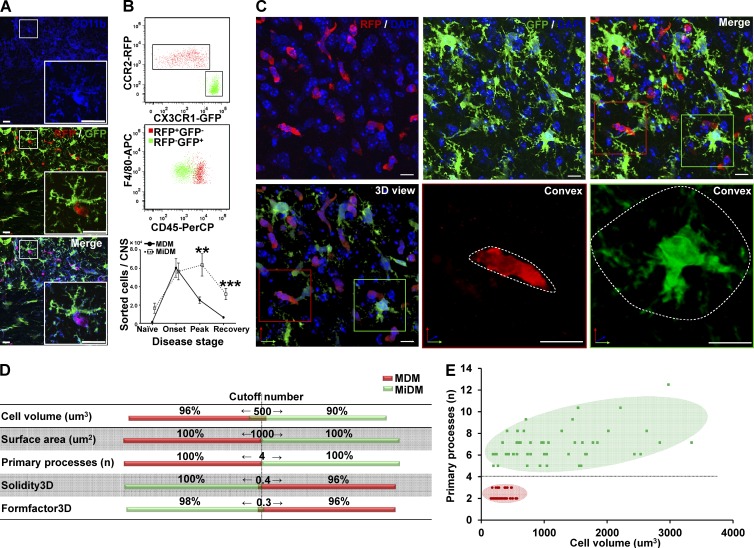
The histological strategy in this study is shown in Table 1. At onset of EAE, two pools of CD11b+ mononuclear phagocytic cells (putative red MDMs and green MiDMs) predominated in spinal cord (Fig. 1 A), indicating that fluorochrome markers could be distinguished at this time point. Using cells isolated from _Ccr2_rfp/+::_Cx3cr1_gfp/+ spinal cords at disease onset, flow cytometry demonstrated distinct expression of RFP and GFP by F4/80+/CD45high MDMs and F4/80+/CD45dim MiDMs, respectively (Fig. 1 B). Enumeration of cells recovered from cell sorting using F4/80+RFP+ as MDMs gate and F4/80+GFP+ as MiDMs gate indicated that MDMs and MiDMs showed equal numbers at disease onset when explosive MDM accumulation occurred. MiDM expansion began at peak (Fig. 1 B). At recovery, MiDMs were found near preonset numbers as MDM frequency continued to decline, which is compatible with previous studies (Ajami et al., 2011). Therefore, there were unequal numbers of MiDMs/MDMs before and after disease onset (Fig. 1 B). Morphological analyses and definitions of relations between myeloid cells and axoglial units were conducted at disease onset so that equal numbers of MDMs and MiDMs could be assayed and early events in the demyelinating disorder could be explored.
Table 1.
Histology analysis strategy
| Method | Purpose | Finding |
|---|---|---|
| Confocal analysis of 0.2 mm optical sections (n = 104 cells). | To distinguish MDMs (RFP+) from MiDMs (GFP+). | MDMs and MiDMs can be distinguished by cell volume and primary processes. |
| SBF-SEM inspection in 0.2 mm sections from 14 lesions, 7 mice at EAE onset. | To detect MDMs and MiDMs in SBF-SEM using cell volume and process criteria. | Using criteria detected in the previous step, it is possible to distinguish MDMs and MiDMs in SBF-SEM images. |
| SBF-SEM inspection of ultrastructure of MDMs and MiDMs. | To detect ultrastructural characteristics of MDMs and MiDMs. | MDMs and MiDMs show characteristic ultrastructural differences regarding their mitochondria, nuclei, osmiophilic granules and microvilli. |
| Quantification of relation of MDMs (n = 169) and MiDMs (n = 86) to axoglial units (n = 29 intact axons, 46 demyelinated axons). | To determine relationship of MDMs and MiDMs to axoglial units and characterize presence of myelin debris. | Most (55/75; 73%) axoglial units are contacted in limited fashion by MDMs and MiDMs. If one cell type is present (20/75 cells), it’s nearly always (18/20 segments) MDMs. |
| Reconstruction of 3D shape of four representative MDMs at axoglial units. | To detect relationship of MDMs with axoglial units at EAE onset. | In all, 49 MDMs interacting with axoglial units in absence of nearby MiDMs, 2-3 MDMs were attached to each (n = 18) axoglial unit. MDMs have close relationship with nodes of Ranvier (7 MDMs /75 axoglial units). 3D reconstructions showed four representative MDMs at axoglial units: show one carrying out active demyelination, three at nodes of Ranvier. |
Figure 1.
MDMs and MiDMs exhibit different time courses of accumulation in the CNS of mice with EAE and morphological characteristics can distinguish them. (A) Immunohistochemistry shows expression of CD11b for red RFP+ MDMs and green GFP+ MiDMs in the spinal cords of _Cx3cr1_gfp/+::_Ccr2_rfp/+ mice at EAE onset. Bars: 25 µm. We studied 6 mice at EAE onset from 3 EAE inductions. In each EAE induction, 8–10 mice were used and 2 mice were selected from each induction. (B) Flow cytometric analysis of CCR2-RFP+ and CX3CR1-GFP+ populations in cells gated for F4/80 expression (top); CD45 expression of F4/80+RFP+ MDMs and F4/80+GFP+ MiDMs populations (middle); and MDMs and MiDMs numbers at EAE onset, peak, and recovery (bottom). We studied 3 mice for naive groups; 12 for onset; 15 for peak; 13 for recovery from 5 EAE inductions. For each induction, 8–10 mice were used and 2–3 mice were selected at each time point (onset, peak, and recovery) for analysis. (C) Confocal microscopy assessment of myeloid cell morphology in lumbar spinal cord from mice at EAE onset. We studied 54 MDMs and 51 MiDMs of 5 EAE onset mice from 3 EAE inductions for (C–E); 2 sections/mouse; 4–6 cells/section; 8–12 cells /mouse. In each EAE induction, 8–10 mice were induced and 1–2 EAE onset mice were selected from each experiment. Bars, 25 µm. (D) Cell volumes of 500 µm3; surface areas of 1,000 µm2; primary process numbers ≤3 or ≥5; solidity3D of 0.4; and Formfactor3D of 0.3 discriminate between MDMs and MiDMs. (E) Model plot of cell volume against primary process number to distinguish MDMs (red symbols and pink area) from MiDMs (green symbols and green area).
Morphological features distinguish MDMs and MiDMs at EAE onset
Immunofluorescence staining for RFP and GFP in spinal cord at EAE onset showed that red MDMs exhibited elongated or spindle shape, whereas green MiDMs showed a process-bearing morphology (Fig. 1 C). Quantification in 3D reconstructions from 0.2-µm confocal z-stack images showed that MiDMs exhibited much larger size than MDMs along with multiple primary processes, which were sparse in MDMs (Fig. 1 D). Several 3D shape parameters also discriminated between MDMs and MiDMs (Fig. 1 D). We observed scant overlap of several values between MDMs and MiDMs (Fig. 1 D), and entirely nonoverlapping distributions for cell volume and primary processes (Fig. 1 E).
MDMs and MiDMs exhibit differentiating ultrastructural characteristics at EAE onset
We used confocal microscopy in 0.2-µm optical sections to correlate structural features of MDMs and MiDMs with RFP or GFP fluorescence, as a bridge to characterizing cells in 0.2 µm SBF-SEM images (Table 1). Using this approach (Fig. 1 E), MDMs and MiDMs were identified by estimating volume and counting primary processes. Volume estimations came from multiplying the midcell area by the number of sections in which the cell was identified. In electromagnetic (EM) images, quantitative analysis also demonstrated differentiating ultrastructural characteristics for mitochondria, nuclei, cytoplasmic osmiophilic granules and microvilli (unpublished data). MDMs had shorter, thicker mitochondria than MiDMs (unpublished data). Total mitochondrial numbers and volumes were equal in MDMs and MiDMs (unpublished data). MDMs had bilobulated or irregular nuclei, whereas MiDMs had round nuclei (unpublished data). MDMs, but not MiDMs, frequently contained osmiophilic granules and microvilli (unpublished data). Collectively, these ultrastructural features provided confirmatory ultrastructural characteristics to distinguish MDMs from MiDMs.
MDMs initiated demyelination at EAE onset
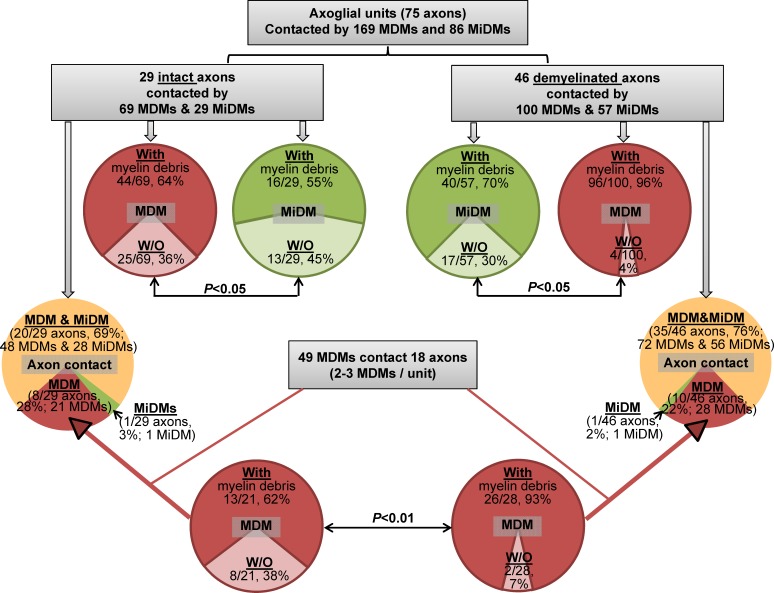
Results from confocal and EM analysis yielded a secure basis for examining the relationships of MDMs and MiDMs to axoglial units at EAE onset (n = 7 mice; 14 lesions) using serial block-face scanning electron microscopy (SBF-SEM), as presented diagrammatically (Table 1). We quantified contacts made by MDMs (n = 169) and MiDMs (n = 86) with axoglial units (n = 75; Fig. 2), and observed that most (55/75; 73%) of all segments (both intact and demyelinated) contacted both MDMs and MiDMs (Fig. 2). Where only one myeloid cell type was present (20/75; 27%), nearly all axoglial units made contacts to MDMs (Fig. 2). In particular, 8/29 intact and 10/46 demyelinated axoglial units were contacted solely by MDMs. We found 2–3 MDMs attached to each of the 18/20 (90%) axoglial units where only MDMs were present (Fig. 2). More than half of all analyzed MDM and MiDM cells (n = 255 total) contained myelin debris, regardless of whether axon segments were intact or demyelinated (Fig. 2). Of the MDMs found in sole contact with axoglial units, virtually all (>90%) MDMs contained myelin when found in sole contact with a demyelinated axon (Fig. 2). These findings motivated evaluation of relationships of MDMs to axoglial units by 3D reconstruction of SBF-SEM image stacks.
Figure 2.
SBF-SEM shows MDMs initiating demyelination at EAE onset. Quantitation of MiDMs and MDMs interacting with axoglial units in SBF-SEM images of CNS at EAE onset. Intact (69%) and demyelinated (76%) segments interacted with MDMs and MiDMs. Red and pink, MDMs; green and light green, MiDMs; yellow, both MDMs and MiDMs. We studied 29 intact axon segments, 46 demyelinated axon segments, 86 MiDMs, and 169 MDMs in 14 lesions of 7 EAE onset mice from 3 EAE inductions as follows: 8–10 mice were immunized at each experiment and 2–3 onset mice were selected from each induction.
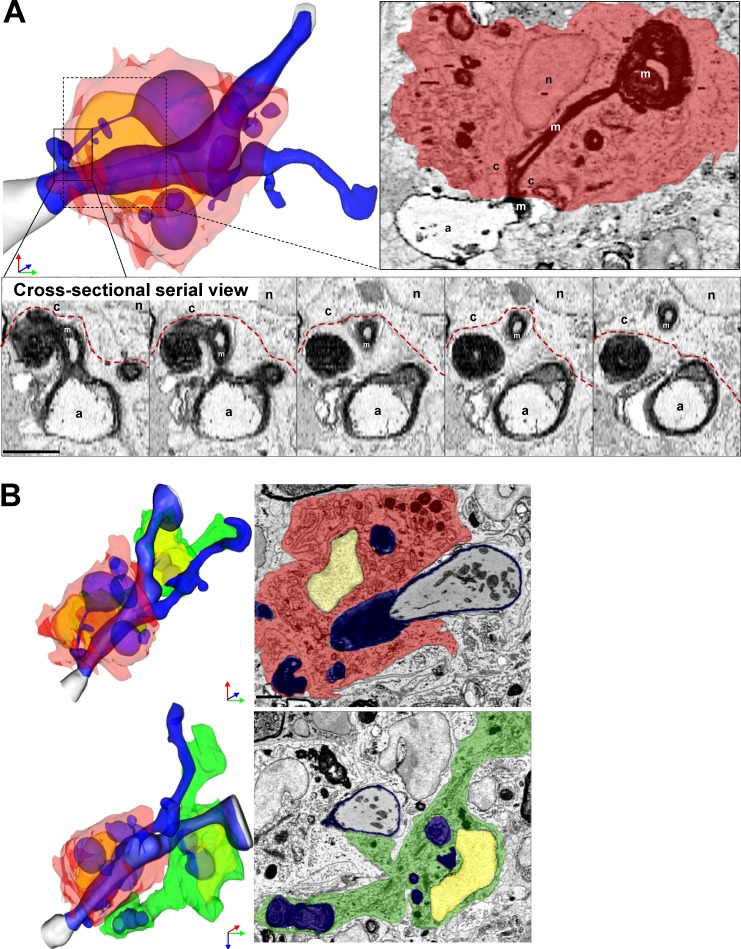
MDMs frequently exhibited morphological characteristics suggesting an involvement in active demyelination. Reconstruction of one representative image stack shows MDMs with large intracellular myelin inclusions tightly encircling a partially demyelinated axon (Fig. 3 A). The myelin peeled away from the axon remained in continuity with a large myelin inclusion inside the MDMs (Fig. 3 A). Remaining myelin was undergoing vesicular breakdown (Fig. 3 A). In contrast, a nearby MiDM encompassed a large fragment of myelin debris (Fig. 3 B) and contacted the nearby MDMs (Fig. 3 B), but made minimal connection to the axoglial unit (Fig. 3 B). In our SBF-SEM data, only MDMs seemed to be implicated in active damage to myelin. These observations suggested that MDMs initiated demyelination at the onset of EAE.
Figure 3.
SBF-SEM shows example of MDM-initiating demyelination at EAE onset. (A) Representative MDMs encircles the axoglial unit. A myelin ovoid within an intracellular phagolysosome shows physical continuity with myelin remaining attached to an axoglial unit which is undergoing active demyelination. In serial images, disrupted myelin shows continuity from outside to inside the MDM. (B) Rotated view from B demonstrating MDM-extensive attachment to axoglial unit and MiDM nearby with limited attachment to axon. A, axon; m, myelin; c, cytosol; n, nucleus. Red, MDM cytosol; green, MiDM cytosol; yellow: nuclei; blue, myelin and myelin debris; gray, axoplasm; red line, MDM plasma membrane. We studied 14 lesions from 7 EAE onset mice from 3 EAE inductions as follows: 8–10 mice were immunized at each experiment and 2–3 EAE onset mice were selected from each induction. Bar, 2 µm.
MDMs surrounded apposed and invaded nodes of Ranvier at EAE onset
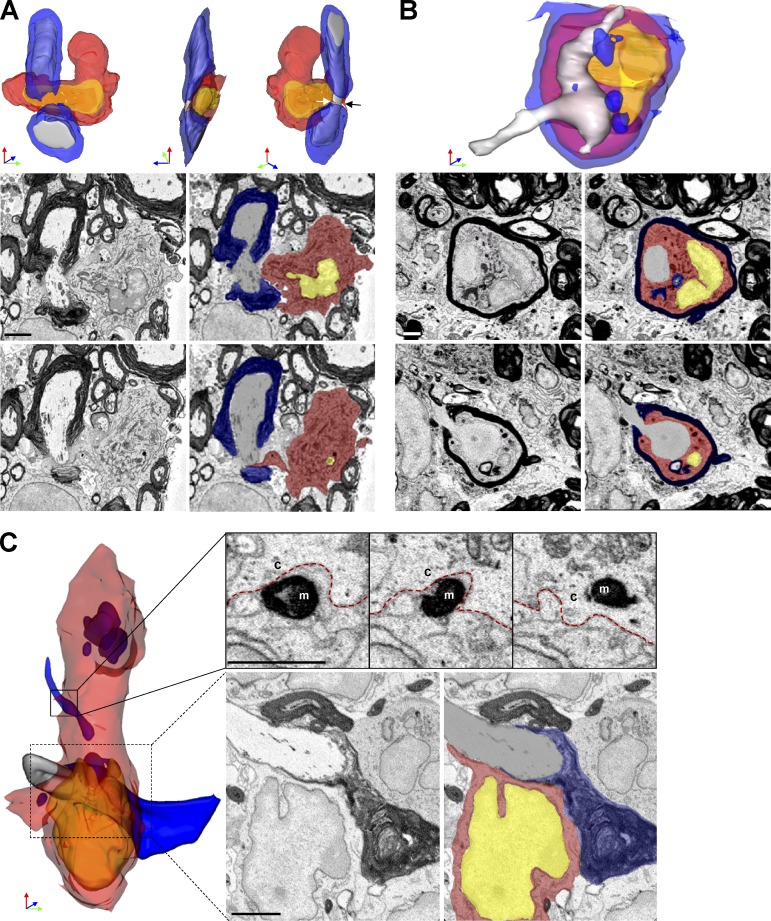
We analyzed axoglial units to examine the nature of contacts with myeloid cells. Unexpectedly, 7/75 (9%) of axoglial units demonstrated MDMs attached to nodes of Ranvier. In each case, the contact between MDMs and node appeared to be pathogenic. One representative monocyte surrounded a node of Ranvier with two microvilli interposed between myelin and axolemma near the paranode complex (Fig. 4 A). The axoglial unit appeared otherwise healthy and no myelin debris was found in the MDM cytosol. This observation suggested that initial MDM–axoglial contacts might occur at nodes of Ranvier. Further, we detected an intratubal (Stoll et al., 1989) MDMs with myelin debris interposed between compact myelin and axolemma near a node of Ranvier (Fig. 4 B). Additionally we identified an MDM apposed to a node of Ranvier and actively phagocytizing myelin (Fig. 4 C). At this node, paranode loops were disrupted and surrounded by MDM cytosol (Fig. 4 C), indicating likely involvement in damaging myelin near the node. No MiDMs contacted nodes of Ranvier.
Figure 4.
MDMs surrounded, apposed, and invaded nodes of Ranvier at EAE onset. (A) SBF-SEM images and 3D reconstruction of SBF-SEM images of MDMs with a node of Ranvier. White and black arrow: microvillus. (B) SBF-SEM images and 3D construction of intratubal MDMs with demyelinated axon and node of Ranvier. (C) SBF-SEM images and 3D reconstruction of an MDM with intracellular myelin debris apposed to a node of Ranvier. Red, MDM cytosol; yellow, nucleus; blue, myelin; gray, axoplasm. M, myelin; c, cytosol. red line, MDM plasma membrane. We studied 14 lesions from 7 EAE onset mice collected as follows: 8–10 mice were immunized at each induction and 2–3 EAE onset mice were selected from each immunization. Bar, 2 µm.
Nodal pathology without demyelination at EAE onset in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice
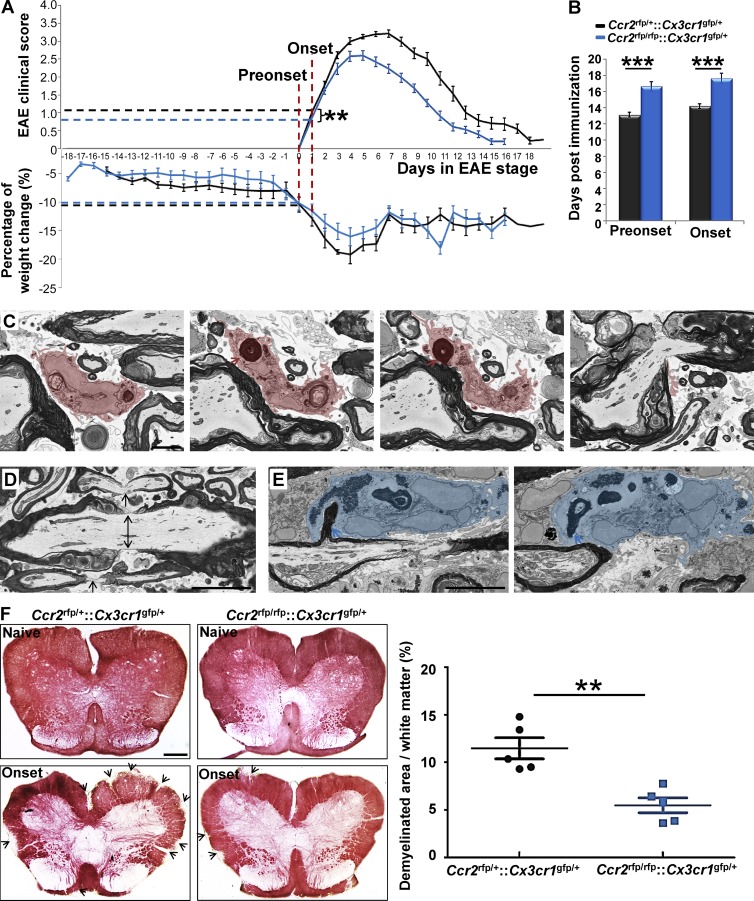
We interpreted our ultrastructural findings to indicate that MDMs recognized altered nodal structure and initiated demyelination at EAE onset. CCR2 is essential for monocyte recruitment to CNS tissues during immune-mediated inflammation (Fife et al., 2000; Izikson et al., 2000; Savarin et al., 2010). To address the role of MDMs in demyelination at EAE onset, we investigated clinical characteristics in relation to node pathology and demyelination in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice in which MDMs were virtually absent from inflamed EAE tissues and replaced in large part by neutrophils (Saederup et al., 2010). We observed equivalent magnitude of weight loss in _Ccr2_rfp/+::_Cx3cr1_gfp/+ and _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice at preonset and onset stages of EAE, showing that CCR2 deficiency did not affect systemic inflammation in this model (Fig. 5 A). There was a moderate delay in disease onset (Fig. 5 B) and slight reduction in EAE onset severity (Fig. 5 A) in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice.
Figure 5.
Nodal pathology without demyelination at EAE onset in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice. (A) Magnitude of weight loss in _Ccr2_rfp/+::_Cx3cr1_gfp/+ and _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice at preonset and onset stages of EAE. Clinical score in _Ccr2_rfp/+::_Cx3cr1_gfp/+ and _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice at EAE onset stage. (B) Days at disease preonset and onset stages of EAE. We studied 28 _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice and 26 _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice for A and B. Data were collected from 12 EAE inductions in _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice and 19 EAE inductions in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice as follows: 8–10 mice were immunized at each induction and 1–3 EAE recovery mice were selected from each immunization. **, P < 0.01; ***, P < 0.001. (C) SBF-SEM imaging of MDMs with nodes of Ranvier phagocytosis in _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice at EAE preonset. Pink, MDM cytosol; red arrow, myelin inclusion of MDM connecting to the node of Ranvier. We studied 3 tissues from 3 _Ccr2_rfp/+::_Cx3cr1_gfp/+ EAE mice at preonset stage from 3 EAE inductions: 8–10 mice were immunized at each experiment and one EAE preonset mouse was selected from each induction. Bar, 2 µm. (D) SBF-SEM of disrupted nodes (black arrows) in preonset spinal cord tissues of _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice. Bar, 2 µm. (E) SBF-SEM of neutrophil is with myelin phagocytosis from internode at preonset stage of _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mouse. Blue, neutrophil cytosol. For D–E, we studied three tissues from three _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ EAE mice at preonset stage from 3 EAE inductions: 8–10 mice were immunized at each experiment and one EAE preonset mouse was selected from each induction. Bar: 2 µm. (F) Histochemical staining and with aurohalophosphate complexes (black gold staining) and quantification of demyelinated area of _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice and _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice. We studied 5 naive _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice, 5 naive _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice, 5 onset _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice, and 5 onset _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice from 3 EAE inductions as follows: 8–10 mice were immunized at each experiment and 1–2 onset mice were selected from each induction. **, P < 0.01. Bar: 250 µm.
SBF-SEM was used to evaluate nodal pathology, myeloid cell relations to axoglial units and demyelination at and before EAE onset. In three distinct tissues from individual _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice with EAE preonset, we found five MDMs attached to disrupted nodes of Ranvier. In an equivalent sample of EAE tissues from three _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice, only one MDM was found in contact with a node of Ranvier, despite the presence of disrupted nodes in proximity to neutrophils. One representative MDM from _Ccr2_rfp/+::_Cx3cr1_gfp/+ tissue having concave nucleus (Fig. 5 C, left) had multiple intracellular myelin inclusions, one of which (Fig. 5 C, left middle) was physically connected to a myelin sheath (Fig. 5 C, right middle) at a paranode (Fig. 5 C, right), indicating active ongoing demyelination at a node of Ranvier. By distinct contrast, EAE onset tissues of _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice were characterized by nodal pathology often without cellular infiltrates (Fig. 5 D). In one instance, we detected a neutrophil abstracting myelin from the myelin internode (Fig. 5, left and right) despite a nearby disrupted node (Fig. 5, left) in tissues from a _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mouse. Importantly, there was no evidence for neutrophil recognition of disrupted nodes of Ranvier. We interpreted these observations to suggest that MDMs specifically recognized nodal components to initiate demyelination, and that absence of MDMs at disrupted nodes of _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice with EAE was caused by the virtual absence of infiltrating monocytes (Saederup et al., 2010).
To quantify the outcome of these ultrastructural differences, we monitored demyelination using histochemical staining with aurohalophosphate complexes at disease onset in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ and _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice. Demyelination was significantly reduced at EAE onset in CCR2-deficient mice (Fig. 5 F), indicating the importance of MDM recognition of disrupted nodes for efficient inflammatory demyelination. Furthermore, as nodal pathology was equivalent in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ (Fig. 5 D) and _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice at the preonset stage of EAE, the results suggested that inflammatory nodal disruption could be reversible if MDMs were prevented from initiating demyelination at those sites.
Expression profiling demonstrates differential MiDMs and MDMs gene expression across the time course of an EAE attack
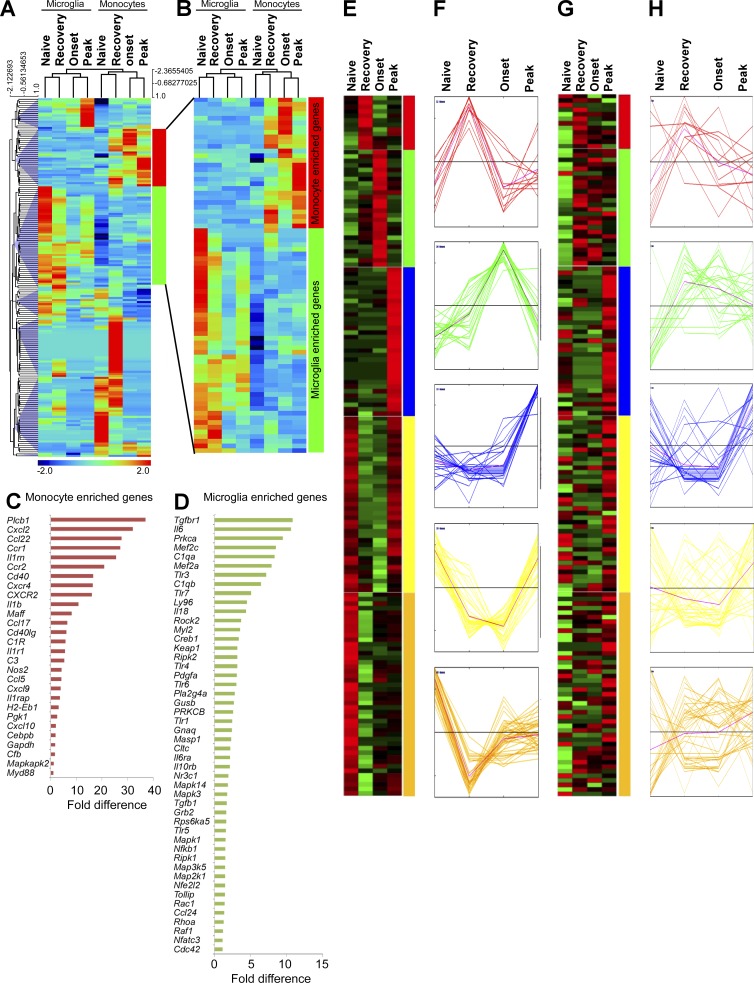
We reasoned that different phenotypes (Fig. 1) and effector properties (Figs. 2–4) of MDMs and MiDMs should be reflected in distinct gene expression profiles in the dynamic CNS microenvironment during EAE. To address this hypothesis, nCounter digital multiplexed gene expression analysis (Kulkarni, 2011) was performed using directly ex vivo naive microglia and splenic F4/80+ macrophages (here termed monocytes and considered similar to microglia by expression profiling; Gautier et al., 2012), as well as flow-sorted MiDMs or MDMs across the time course of an EAE attack. Microglia and MiDMs clustered together during unsupervised hierarchical clustering, as did monocytes and MDMs (Fig. 6, A and B). In both MiDMs and MDMs, naive and recovery-stage expression profiles were more alike than were onset and peak-stage profiles (Fig. 6, A and B) suggesting a return to homeostasis at EAE recovery. We noted a subset of genes that were expressed in microglia and highly regulated in MiDMs during EAE, but not expressed at all in monocytes or MDMs (Fig. 6, A and B). Conversely, a subset of MDM-enriched genes were dynamically regulated in monocytes and MDMs but not in microglia (Fig. 6, A and B). MDM-enriched genes were sharply up-regulated from naive monocytes to onset and peak-stage MDMs (Fig. 6 B), descending toward naive levels during recovery (Fig. 6 B). In contrast, MiDM-enriched genes were strongly expressed in naive cells, almost uniformly silenced at onset, and began a return toward naive levels at peak and recovery (Fig. 6 B). Comparing MDM-enriched genes with MiDM-enriched genes showed that MDMs were more likely to express effector functions, including secreted factors and surface molecules (18/28; 64.3% of MDM-enriched genes encoded effector functions; Fig. 6 C; and Table S1, purple genes). In contrast, only 18/48 (37.5%) of MiDM-enriched genes encoded effector functions (Fig. 6 D, Table S1, purple genes). These observations indicated that MiDMs and MDMs exhibited markedly distinct expression profiles during EAE.
Figure 6.
Inflammatory signature in MiDMs versus MDMs in the CNS of _Cx3cr1_gfp/+::_Ccr2_rfp/+ mice with EAE. (A) Quantitative nCounter expression profiling of 179 inflammation related genes was performed in CNS-derived GFP+ microglia and RFP+ recruited monocytes from naive and EAE mice at onset, peak and recovery stages. Each row of the heat map represents an individual gene and each column an individual group from pool of 5 mice at each time point. The relative abundance of transcripts is indicated by a color (red, higher; green, median; blue, low). For A–H, we studied five mice in each time point (onset, peak, recovery) from 3 EAE inductions; 8–10 mice were immunized in each induction. (B) Heat map of differentially expressed microglia and monocyte genes. (C) Enriched monocyte genes as compared with resident microglia. Bars represent fold changes of gene expression across naive and all disease stages versus resident microglia. (D) Enriched microglia genes as compared with recruited monocytes. Bars represent fold changes of gene expression across naive and all disease stages versus recruited monocytes. (E–H) K-means clustering of inflammation genes in resident microglia and recruited monocytes. K-means clustering was used to generate 5 disease stage–related clusters in MiDMs. Heat map (E) and expression profile (F) of inflammation genes in MiDMs are shown by generated clusters. MDM expression matrix overlaid on microglial based clusters shows (G) heat map and (H) expression profile.
Differential expression of macrophage effector functions by MiDMs and MDMs
Our ultrastructural analysis of myeloid cells in EAE focused on myeloid cell relationships to tissue elements. Expression profiling also addressed the cytokine and growth factor output of MiDMs and MDMs, potentially providing insight into disease pathogenesis. We used k-means clustering to discriminate five distinct patterns of MiDM gene expression during the course of EAE (Fig. 6, E and F). The red, blue and green groups increased in MiDMs at onset, peak, and recovery, respectively. Red group genes involved several surface molecules. Green group genes, up-regulated at onset and transiently further up-regulated at peak, were comprised mainly of complement-system elements (C3aR1; C4a, C1qa, C1qa, C3, and Cfb); mononuclear cell–specific chemokines (CCl2, 3, 4, 5, 7, and CXCL9); proliferation related genes (fos, jun, myc, and CSF1); and acute inflammation–related genes (IL1a, IL1b, TNF, CEBP, STAT1). Cell growth–related genes expressed at this time point correlated to reported patterns of microglial proliferation during EAE (Ajami et al., 2011). Blue group genes up-regulated at recovery included heterogeneous cytokines (_IFN_-α, _IFN_-β, TGFB3, IL2, IL3, IL4, _IL12_α, _IL12_β, PDGFA, CSF2, and CXCL2). Both yellow group and golden group genes were strongly expressed in naive microglia, reduced drastically at onset, and either returned to preEAE levels during recovery (yellow) or failed to do so (golden). These genes included a large spectrum of intracellular signaling components from the MAP-kinase pathways, as well as _TGF_β and receptor, both of which are implicated in the naïve microglial phenotype (Butovsky et al., 2014).
These five gene groups were also analyzed for MDM expression patterns during EAE (Fig. 6, G and H). None of the gene groups showed coordinate regulation patterns in MDMs, as were observed in MiDMs (Fig. 6 H). This observation underscored disparate responses of MiDMs and MDMs to the inflammatory CNS microenvironment of EAE, despite their being present in close proximity (Fig. 1 A).
Expression patterns at EAE onset in relation to MiDM and MDM function
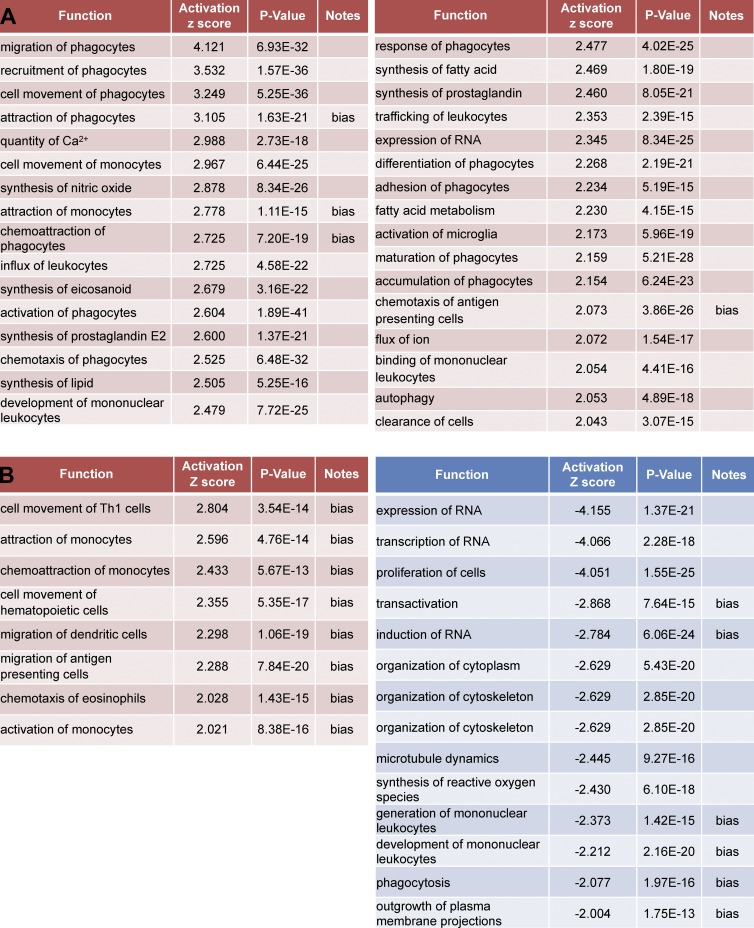
To determine whether gene expression patterns could be informative for understanding the relationships of cells to axoglial elements in tissues at EAE onset, we interrogated naive versus onset MDM and MiDM gene expression related to cellular functions (Fig. 7). MiDMs showed highly significant up-regulation of functions associated with cell movement, chemoattraction, and migration (Fig. 7 B). In the Ingenuity IPA database, the terms cell movement, chemoattraction, and migration indicated production of chemokines such as CCL2, CCL3, CCL4, CCL5, and CCL7, which are up-regulated at onset and further increased at peak (Fig. 6, E and F, green group and genes). In other respects, MiDMs exhibited a repressed metabolic and activation phenotype by comparison to naive microglia (Fig. 7 B) including proliferation, RNA metabolism, cytoskeletal organization, microtubule dynamics, extension of processes, phagocytosis and generation of reactive oxygen species.
Figure 7.
Affected functions in MiDMs and MDMs at EAE onset. nCounter inflammatory gene expression data were uploaded to IPA. Genes with fold change (EAE onset vs. Naive) ≥1.5 or ≤−1.5 were included in downstream effects analysis. (A) MDMs up-regulated functions, sorted by activation z-score. (B) MiDMs up-regulated (left) and down-regulated (right) functions, sorted by activation z score. The bias term indicates imbalanced numbers of up- and down-regulated genes associated with a distal function requiring significance at the P < 0.01 level. We studied pooled samples from 5 mice in each time point (onset, peak, recovery) from 3 EAE inductions; 8–10 mice were immunized in each induction.
MDMs showed up-regulation of functions associated to macrophages, including phagocytosis, calcium signaling, production of prostanoids, adhesion, autophagy, and cell clearance (Fig. 7 B). This pathway analysis corresponded well to effector properties displayed by MDMs in our SBF-SEM analysis (Figs. 2–4). No functions were reported to be down-regulated in MDMs at EAE onset as compared with naive monocytes.
A comprehensive listing (Table S1) of all genes regulated by at least twofold in MiDMs or MDMs as compared with expression levels in naive mice affirmed and extended these interpretations. At EAE onset when SBF-SEM analyses were performed, MiDMs predominantly suppressed the distinctive gene expression pattern which correlates to their unique phenotype (Chiu et al., 2013), reflected by the observation that MiDMs down-regulated far more genes than were up-regulated (Table S1). In contrast, MDMs up-regulated far more genes than did MiDMs and up-regulated more transcripts than were down-regulated. Additionally, the extent of gene up-regulation in MDMs exceeded that seen in MiDMs.
MiDM and MDM gene expression kinetics reflected return toward homeostasis in recovery stage
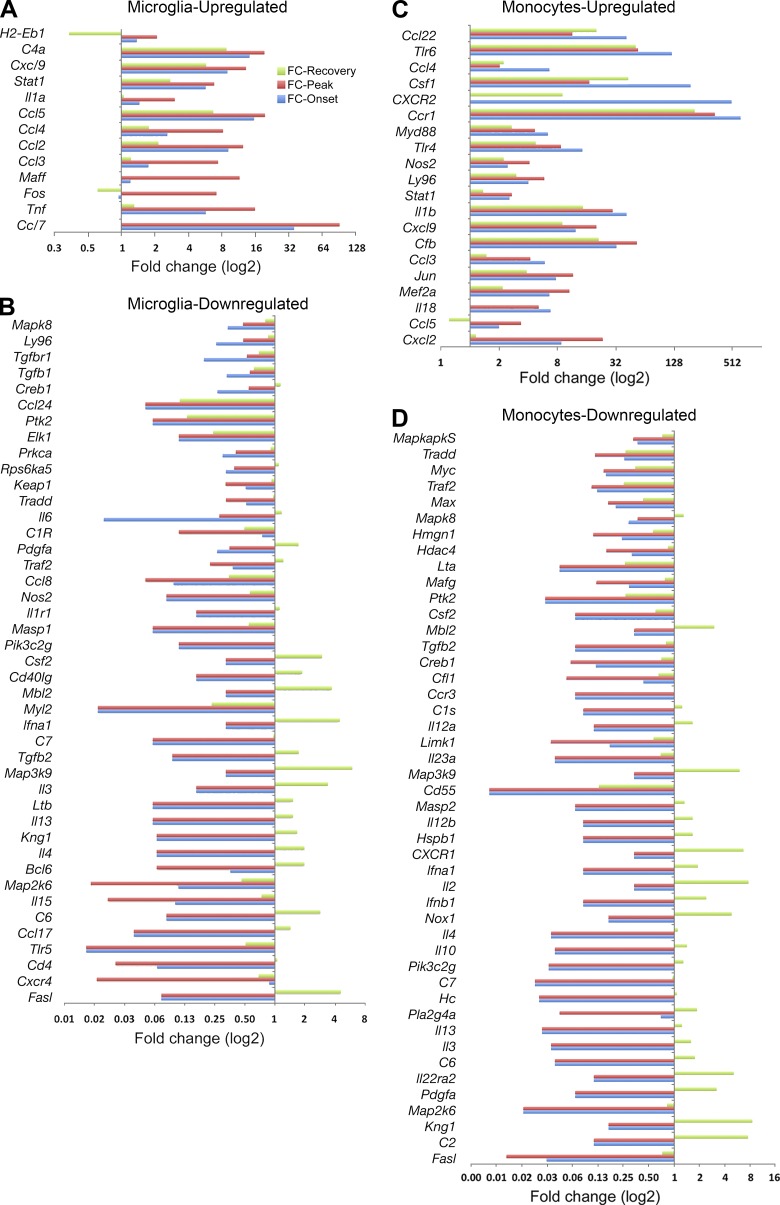
These expression profiles showed consonant changes for the vast majority of genes analyzed: if a gene was up-regulated at any time point, then its expression level showed an increase at other time points as well. However, a substantial minority of genes both for MDMs and MiDMs showed some dissonant time points, at which a previously down-regulated gene might show up-regulation (unpublished data). We show this subset of recovered genes in Fig. 8. In virtually every case (Fig. 8, A–D), these dissonant compensatory changes took place during recovery and almost always showed an increase in a gene that had been down-regulated during onset and peak. Both MiDMs (Fig. 8 B) and MDMs (Fig. 8 D) demonstrated this pattern of gene-expression kinetics.
Figure 8.
Restoration of affected inflammatory genes in resident microglia and recruited monocytes at recovery stage. For each gene, fold change of all different disease stages versus naive state were calculated. Genes that contained at least one fold change >2 or < 0.5 and average fold change for peak and onset >2 or <0.5 were presented. (A) Microglial up-regulated; (B) Microglial down-regulated; (C) monocyte up-regulated; (D) monocyte down-regulated. We studied pooled samples from five mice in each time point (onset, peak, recovery) from three EAE inductions; 8–10 mice were immunized in each induction. FC, fold change.
Convergent and divergent responses to upstream regulatory signaling by MiDMs and MDMs
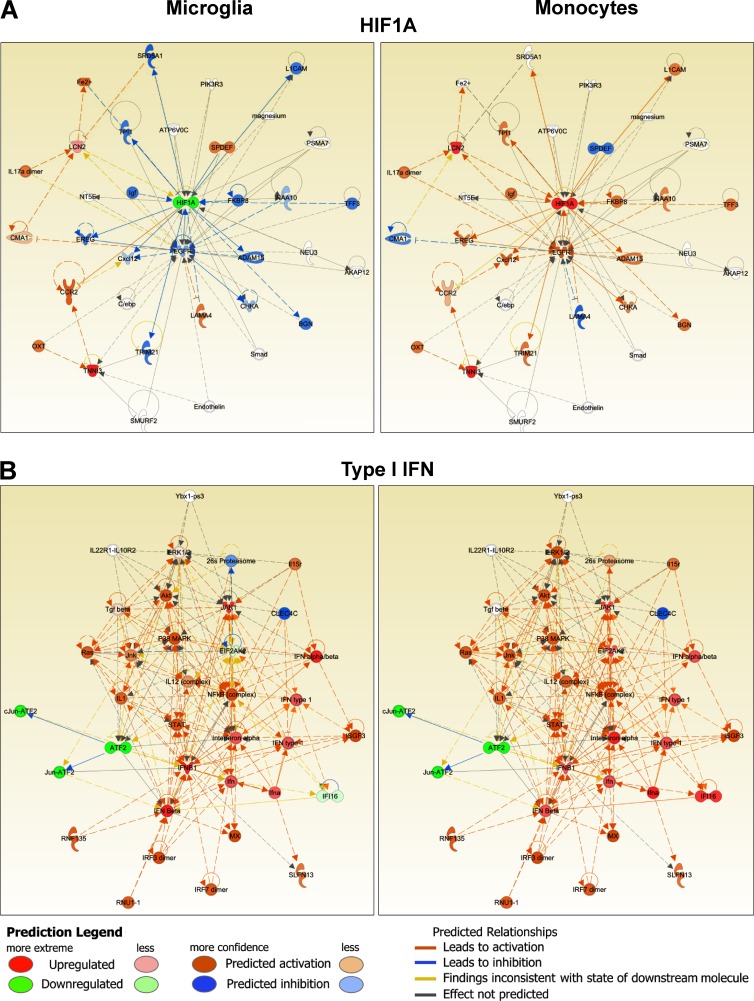
Translation of observations made using expression profiles can be enabled through identification of upstream regulators. We used Ingenuity IPA software to identify putative upstream regulators of the gene expression alterations demonstrated by MiDMs and MDMs at disease onset. Putative regulatory elements were then grouped in signaling modules and subjected to pathway analysis. Cell motility pathways were clearly different in MiDMs and MDMs (unpublished data). Core elements such as RhoA (Xu et al., 2009) were regulated divergently and associated signaling components were predicted to be enhanced in MDMs but depressed in MiDMs, consistent with our phenotypic characterization using SBF-SEM. Both HIF-1α (Fig. 9 A) and TNF pathways (not depicted) were also differentially regulated in MiDMs and MDMs. By contrast, type I IFN pathway (Fig. 9 B) was regulated virtually identically in MiDMs and MDMs. Collectively, these data suggest that HIF-1α and TNF signaling may partly drive pathogenic properties of MDMs. Additionally, these data indicated that the separate ontogeny of microglia and monocytes will lead, probably by epigenetic influences, to divergent responses to some but not all environmental stimuli, with phenotypic consequences according to the CNS microenvironment.
Figure 9.
Function networks in MiDMs and MDMs. nCounter inflammatory gene expression data were uploaded to IPA. Genes with fold change (EAE onset vs. Naive) ≥1.5 or ≤−1.5 were included in upstream regulators analysis. Predicted upstream regulators were manually curated to form functional clusters. Clusters were uploaded to IPA using Z scores as reference value for each gene. Networks were generated for each cluster consisting of uploaded genes and additional predicted molecules. (A) Typical example of functions with dissimilar activation pattern in MiDMs and MDMs: HIF1A. (B) Function with similar activation pattern in MiDMs and MDMs: Type I IFN. Red object denotes positive (>2) z score and green object denotes negative (<2) z-score. Orange object denotes predicted activation of the network object. Blue object denotes predicted inhibition of the network object. Predicted relationships (connecting lines): orange, leads to activation; blue, leads to inhibition; yellow, finding inconsistent with state of downstream molecule; gray, effect not predicted.
DISCUSSION
In this study, we developed a novel strategy to discriminate MDMs from MiDMs. We used SBF-SEM to address the detailed relationships of MiDM and MDM to axoglial units in the spinal cords of mice at EAE onset and expression profiling to examine potential mechanisms. Selection of the EAE disease model ensured that both recruited monocytes and resident microglia were exposed to the same intensely inflammatory environment to increase the likelihood that ambient conditions could activate these two myeloid cell types toward a convergent inflammatory phenotype. Instead, we found strikingly divergent relationships of MDMs and MiDMs to axoglial units, by quantitative and qualitative ultrastructural analysis. Results from expression profiling supported this interpretation by showing that MiDM metabolism was severely down-regulated, whereas expression profiles of MDMs reflected the activated phagocytic phenotype observed through SBF-SEM.
Several salient new observations emerged from these experiments. First, we showed that MDMs initiate demyelination at EAE onset, as MDMs were the overwhelmingly dominant cells found in isolation attached to axoglial units and demonstrated destructive interactions with myelinated axons in 3D reconstructions. Second, MDMs were unexpectedly observed at nodes of Ranvier in 9% of axoglial units and showed remarkably invasive behavior, including extension of microvilli (Fig. 4 A) or localization of cell soma (Fig. 4 B) between axolemma and myelin sheath. Our observed frequency of MDM–nodal interaction represents a minimum estimate as MDMs found at heminodes adjacent to a demyelinated segment (Fig. 3 B) were not scored. Comparison of _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ and _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice at and before EAE onset emphasized the importance of MDMs for this mechanism of demyelination. In particular, neutrophils in inflamed CNS of _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice did not recognize disrupted nodes. These observations are clinically pertinent: our detection of MDMs at nodes of Ranvier is consistent with recent reports of nodal pathology in clinical demyelinated tissues (Fu et al., 2011; Desmazières et al., 2012). The present observations extend this concept and provide a cellular basis for nodal pathology at the earliest stages of demyelination. Given the presence of potential phagocytic signals at nodes (antibodies to paranodal proteins such as contactin and neurofascins; Meinl et al., 2011); complement-derived opsonins (Nauta et al., 2004); and stress-induced eat-me signals (Hochreiter-Hufford and Ravichandran, 2013), it may be feasible to identify a direct molecular pathway for initiating demyelination in this model. Third, we characterized a molecular signature for resident microglia at EAE onset. Grouping of regulated genes into functional categories demonstrated a remarkable down-regulation of microglial metabolism at the nuclear, cytoplasmic and cytoskeletal levels.
In our initial experiments we found that the presence of myelin debris at the peak of EAE did not discriminate MDMs from MiDMs. We considered that SBF-SEM would exhibit advantages for spatial resolution (Denk and Horstmann, 2004) required for characterizing relationships of myeloid cells to axoglial units during the inflammatory demyelinating process at EAE onset. To take advantage of this technique we developed methods based on cell volume and process number (Fig. 1 E), to distinguish MDMs from MiDMs in 0.2-µm confocal optical sections, and translated this approach directly to SBF-SEM image sets at 0.2-µm intervals. We also noted differential nuclear morphology, mitochondrial shape, and osmiophilic granule content between MDMs and MiDMs. These characteristics of MDMs and MiDMs may not be universally present in other pathological circumstances but demonstrate an approach to ultrastructural distinction of myeloid cell populations in tissue sections.
Gene expression profiling across the time course of EAE yielded intriguing kinetics as analyzed by k-means clustering. Five patterns were observed. Red group genes (increased at onset) comprised the smallest number and involved several surface molecules: CCR1, CCR7, CXCR2, and CD40. Of these, CCR7 and CD40 have been reported on activated microglia, including those observed in MS tissue sections (Kivisäkk et al., 2004; Serafini et al., 2006). GAPDH was up-regulated in MiDMs at onset. Although often regarded as a housekeeping gene, GAPDH is found in complexes that limit the translation of inflammatory gene transcripts in activated mouse macrophages (Mukhopadhyay et al., 2009; Arif et al., 2012). As previously reported (Chiu et al., 2013), MiDM gene expression during the course of EAE did not correspond to the M1/M2 pattern of peripheral macrophage responses to infection or tissue injury. Microglial morphological transformation can be relatively uniform regardless of the inflammatory process that provokes it. Despite this apparent uniformity, gene expression by morphologically identical microglia can differ drastically contingent on context (Perry et al., 2007).
Unsupervised hierarchical clustering provided insight into gene expression patterns of MDMs and MiDMs. Naive and recovery patterns were similar for both cell types. At disease onset, microglia showed drastic down-regulation of the expression profile observed in cells from healthy brain. Brisk microglial proliferation (Ajami et al., 2011) may have accelerated a gradual return toward a homeostatic expression profile, as suggested by MiDM up-regulation of fos, jun, myc, and CSF1 (Wei et al., 2010) at disease onset. By striking contrast, MDMs up-regulated a large suite of inflammation-associated genes at EAE onset, with subsequent regression to the expression phenotype of circulating monocytes.
Blood monocytes and resident microglia were exposed to the same inflammatory environment. However, their preEAE states were extremely distinct, with monocytes being generated from a bone marrow progenitor within weeks of entry into CNS, whereas microglia originated during early embryogenesis and had inhabited a serum-free unique environment from midgestation. In a recent study, we characterized resident microglia by profiling mRNA, miRNA, and protein in comparison with infiltrated brain macrophages, nonmicroglial resident brain cells, and peripheral macrophages (Butovsky et al., 2014). The detailed profiling after separating cells via CD45dim status showed distinct mRNA, miRNA, and protein expression by microglia as compared with infiltrating monocytes or neuroepithelial brain cells (Butovsky et al., 2014). The study described transcription factors and miRNAs characteristic of microglia in healthy brain but not in peripheral monocytes. These findings partially explain a divergent response of these two cell types to the same stimuli (Butovsky et al., 2014).
The strength of the study is that the dual reporter system is sufficient to accurately distinguish monocyte versus microglial cells and thus to address the general concept that monocytes and microglia can exert differential functions in a CNS disease process. At the onset of EAE, the time point at which our imaging studies were focused, we are able to make an unequivocal distinction between resident microglia (CX3CR1gfp) and infiltrating monocytes (CCR2rfp). Two empirical observations underline this discrimination: microglia are uniformly CX3CR1+ from early embryonic time points through adulthood (Cardona et al., 2006; Ginhoux et al., 2010; Schulz et al., 2012), and CCR2+Ly6C+ cells constitute the vast majority of infiltrating monocytes at EAE onset (Saederup et al., 2010; Mizutani et al., 2012).
There were unavoidable limitations of our research; specifically, to address how monocytes and microglia respond to a shared microenvironment, we focused on a single, pathogenically relevant time point: onset of EAE. For this reason, it was beyond the scope of our study to decipher the phenotypic fate of infiltrated monocytes. In peripheral models of inflammation, Ly6Chi/CCR2rfp monocytes down-regulate the reporter over time and show phenotypic evolution. Furthermore, our conclusions should not be generalized beyond the present disease paradigm: in other models, such as spinal cord contusion, the inflammatory infiltrate includes Ly6Clow/CX3CR1gfp monocytes, which are highly pathogenic (Donnelly et al., 2011). Our findings carry biological and medical significance by demonstrating and characterizing differential responses of infiltrating monocytes and resident microglia in a relevant disease model at a prespecified time point, at which point pathogenic events are taking place. Therefore, we focused our analysis on the day of EAE onset rather than subsequent events to challenge our overall hypothesis that infiltrating monocytes versus resident microglia respond very differently to acute inflammatory stimuli.
Activated myeloid cells are the proximate effectors of a bewildering array of acute and chronic disorders (Wynn et al., 2013). The technical and conceptual approach taken in this study may be applicable to other tissues and disease processes. In many pathological conditions, tissues harbor a mixed population of activated resident and recruited monocytes. The therapeutic strategy will differ conclusively based on the specific effector properties of each cell type and the stage of disease. In particular, if monocytes are pathogenic, then their trafficking should be blocked using a peripherally active agent. The optimal application of agents that regulate leukocyte migration and intracellular signaling will be promoted by detailed examination of each individual myeloid population.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were obtained from the National Cancer Institute. _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice were generated by crossbreeding _Ccr2_rfp/rfp::C57BL/6 mice (Saederup et al., 2010) with _Cx3cr1_gfp/gfp::C57BL/6 mice (Jung et al., 2000). _Ccr2_rfp/rfp::_Cx3cr1_gfp/gfp mice were generated by breeding _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice. _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice were generated by crossbreeding _Ccr2_rfp/rfp::C57BL/6 mice with _Ccr2_rfp/rfp::_Cx3cr1_gfp/gfp mice. Animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic following the National Institutes of Health guidelines for animal care.
EAE induction and clinical evaluation.
EAE was induced in _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice and _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice of 24–28 wk of age using myelin-oligodendrocyte-glycoprotein peptide 35–55 (MOG) as previously described (Huang et al., 2006). All mice were weighed and graded daily for clinical stages as previously reported (Saederup et al., 2010). We defined clinical stage of EAE as follows: pre-onset was the day sudden weight loss for 8–10% occurred; onset was the day EAE signs appeared; peak was the second day score didn’t increase after sustained daily worsening; and recovery was the second day score didn’t decrease after a period of sustained daily improvement.
To address our research questions, we integrated flow cytometry, immunohistochemistry with quantitative morphometry, cell sorting for expression profiling, and serial block-face scanning electronic microscopy. In all, we performed 12 EAE immunizations in _Ccr2_rfp/+::_Cx3cr1_gfp/+ mice and 19 immunizations in _Ccr2_rfp/rfp::_Cx3cr1_gfp/+ mice for this project, with 8–10 mice in each immunization. We selected EAE mice at onset, peak or recovery depending on the specific studies underway at that time, with the majority of mice coming from the onset stage of EAE. Each experiment incorporated samples from at least three separate immunizations. Details of mouse numbers and how they were selected for each experiment were included in the figure legends as requested.
Cell isolation and flow cytometry.
Brains and spinal cords were removed and homogenized. Mononuclear cells were separated with a 30%/70% Percoll (GE Healthcare) gradient as previously reported (Pino and Cardona, 2011). Single-cell suspensions from CNS were stained with anti–F4/80-APC (BM8; eBioscience) and anti–CD45-PerCP (30-F11; BioLegend). Cells were either analyzed on a LSR-II (BD) or sorted on a FACSAria II (BD) running Diva6. Data were analyzed with FlowJo software (Tree Star).
Histological and immunohistochemical analysis.
Spinal columns were removed after mice were perfused with 4% paraformaldehyde (PFA). For immunofluorescence assay, free floating sections of the lumbar spinal cord were prepared as previously described (Huang et al., 2006). For immunofluorescence assay, sections were blocked with 10% normal serum for 2 h and stained with primary antibodies at 4°C for 24–48 h. After washing with PBS-T (PBS with 0.1% Triton X-100; Sigma-Aldrich) three times, the sections were incubated with secondary antibodies at room temperature for 2 h and mounted in ProLong Gold antifade reagent (Invitrogen). Antibodies used include rat anti-CD11b (BD), mouse anti-GFP (Abcam), rabbit anti-RFP (Abcam), Alexa Fluor 488 goat anti–mouse IgG (Invitrogen), Alexa Fluor 594 goat anti–rabbit IgG (Invitrogen), and Alexa Fluor 647 goat anti–rat IgG (Invitrogen). Nuclei were labeled by DAPI. Images were collected by confocal laser-scanning microscope (SP5; Leica).
Quantitative 3D morphology.
Quantitative 3D morphology of MDMs and MiDMs was analyzed in confocal images from spinal cord of mice at EAE onset. Free floating sections of the lumbar spinal cord were stained with RFP for MDMs, GFP for MiDMs, and DAPI for nuclei. Stack images were taken at 0.2-µm step size along the z-direction with a 63× objective (numerical aperture [NA] = 1.4) and zoom factor 2. A square (1024×1024 pixels) corresponding to 123 × 123 µm2 was used for the analysis. Cells were 3D reconstructed by ImageJ software and all analyses were performed using ImageJ with 3D Convex Hull plugin. The parameters analyzed include voxel (volumetric pixel), convex voxel, volume, convex volume, surface, and convex surface area. Other calculated parameters were: Solidity3D = volume/convex volume; Convexity3D = convex surface area/surface area; Formfactor3D = 36π×volume2/surface area3.3 The number of primary processes was estimated visually. We included 5 mice, 54 MDMs; 51 MiDMs in this assay with 2 sections/mouse, 4–6 cells/section and 8–12 cells/mouse. Those mice came from three EAE inductions.
SBF-SEM.
Spinal cords were removed after mice were perfusion-fixed using 4% PFA with 1% glutaraldehyde. Lumbar spinal cord sections were made on a vibratome (Leica). Sections were stained with 0.4% OsO4, uranyl acetate and lead aspartate, then embedded in epon resin (Electronic Microscopy Sciences). SBF-SEM images were acquired using a Sigma VP SEM (Carl Zeiss) with 3View (Gatan). Serial image stacks of images at 100-nm steps were obtained by sectioning 48 × 48 × 20 µm3 tissue blocks (length × width × depth) at a resolution of 8192 × 8192 pixels. Image stacks were processed for 3D reconstruction by TrakEM2 in FIJI software (National Institutes of Health). Alternating sections from the same stacked images were chosen to make stacks for 3D reconstructions which matched the 0.2-µm step size used for acquiring confocal stacked images. In SBF-SEM images, we discriminated MDMs and MiDMs using the volume/primary processes model (Fig. 1 E) generated from analyzing confocal images. Quantifications of myeloid-cell spatial relationships to axoglial units, including myelin incorporation, were done in SBF-SEM images.
Quantification of nuclei and mitochondria.
Characterizations of nuclear shapes were conducted in SBF-SEM images. Nuclei were categorized as follows: round, round shape and smooth surface with ratio of length/width ≤1.5; elongated, elongated or oval shape with length/width >1.5, and may have small indentations; Bilobulated: two connected lobes with single intervening large indentation; Irregular: complicated shape with corrugated surface, and may have multiple and variable sizable indentations. Blinded observers (n = 3) scoring the nuclear morphology from SBF-SEM images included a research student, a research fellow and a neuroscientist. Observers were trained on the same nuclear examples in each category and practiced using 20 nuclei comprising all shapes before scoring the nuclei. Kappa test showed good pairwise agreement rates among observers (>0.8) and the data from the neuroscientist are used. Quantifications were done in 3 individual mice from 3 EAE inductions including 28–35 cells from two separate lesions from each mouse in the assay.
Mitochondria of MDMs and MiDMs at EAE onset were reconstructed from SBF-SEM images to 3D images using TrakEM2. 5 MDM and 5 MiDM cells from 3 separate mice at EAE onset (total 10 cells) were included in the assay. Those mice came from 3 EAE inductions. Mitochondria were quantified for length, cross-sectional area, volume and ratio of length/cross-sectional area using Fiji software.
Quantification of demyelination.
Black-gold staining was performed according to a protocol described previously (Liu et al., 2010). In brief, 5 free floating lumbar spinal cord sections were stained in 0.2% black-gold solution at 65°C water bath for 10 min. After staining with black-gold, sections were pictured by 3-CCD video camera interfaced with an Image-Pro Plus Analysis System (Version 4.1.0.0; MediaCybernetics) and analyzed with ImageJ software. Demyelinated areas are those void of black-gold staining. Mean percentage of demyelinated areas in white matter were calculated. We included 5 mice from 3 EAE inductions in this assay.
Statistical analysis of cellular elements.
Statistical analyses were performed using SAS (SAS Institute Inc.), PRISM (GraphPad Software) and SPSS 17.5 (SPSS Inc.). Flow cytometry data were analyzed by two-way ANOVA test and Wilcoxon matched-pairs signed rank test. Nuclear shape quantifications were compared by paired Student’s t tests and logistic regression. Mitochondrial quantifications were compared by Mann-Whitney U test and linear mixed model. Quantitative relationships of myeloid cells to axoglial units were compared using logistic regression with generalized estimating equations (GEE). Clinical characteristics of EAE mice were analyzed using two-way ANOVA test with Bonferroni post test. Percentage of demyelination was compared by Student’s t test. Data were shown as mean ± SEM or median (the first quartile–the third quartile) and P < 0.05 was considered statistically significant.
Gene expression.
Mononuclear cells were prepared from brains and spinal cords as described previously. Cells were sorted on a BD FACSAria II by gating on F4/80+GFP+ for MiDMs and F4/80+RFP+ for MDMs. RNA was isolated from FACS-sorted cells mixed from three mice from six EAE inductions per data point in TRIzol Reagent (Ambion) according to manufacturer’s protocol. RNA samples were analyzed by nCounter gene expression analysis and quantified with the nCounter Digital Analyzer (NanoString Technologies). Expressions of 179 genes were analyzed using nCounter GX Mouse Inflammation kit.
Nanostring data normalization.
Normalization was conducted with nSolver Analysis Software1.1. Data were normalized using positive and negative controls and housekeeping genes probes. Background level was calculated for each sample as mean of negative control probes + (x2 SD). Calculated background was subtracted from each gene expression value. In cases where the calculated value was <1, values were set to 1.
Hierarchical and k-means clustering analysis.
Hierarchical cluster analysis was performed using Pearson correlation for distance measure algorithm to identify samples with similar patterns of gene expression. MiDM samples expression data were used in k-means clustering using Pearson correlation for distance measures (Multi Experiment Viewer v. 4.8).
IPA (Ingenuity) analysis.
Data were analyzed using IPA (Ingenuity Systems). Differentially expressed genes (EAE onset MiDMs versus naive microglia and EAE onset MDMs versus naive splenic monocytes) were used in downstream effects and upstream regulators analyses. Uploaded dataset for analysis were filtered using cutoff definition of 1.5-fold change. Level of confidence for analysis was set to high-predicted and experimentally observed.
Terms used in IPA analyses.
The p-value is a measure of the likelihood that the association between a set of genes in the uploaded dataset and a related function or upstream regulator is due to random association. The smaller the p-value, the less likely it is that the association is random and the more significant the association. In general, P < 0.05 indicate a statistically significant, nonrandom association. The p value of overlap is calculated by the Fisher’s Exact Test.
The activation z-score is a value calculated by the IPA z-score algorithm. The z-score predicts the direction of change for a function or the activation state of the upstream regulator using the uploaded gene expression pattern (upstream to the function and downstream to an upstream regulator). An absolute z-score of ≥2 is considered significant. A function is increased/upstream regulator is activated if the z-score is ≥2. A function is decreased/upstream regulator is inhibited if the z-score ≤-2.
The bias term is the product of the dataset bias and the bias of target molecules involved in a particular function annotation or upstream regulator activity. A biased dataset is one where there is more up- than down-regulated genes or vice versa. The dataset bias is constant for any given analysis and the function/upstream regulator bias is unique for each upstream regulator/function. When the absolute value of this term is 0.25 or higher, then that function/upstream regulator’s prediction is considered to be biased and the Fisher’s exact p-value must be 0.01 or lower for the analysis to be considered significant.
Supplementary Material
Supplemental Material
Acknowledgments
We thank Dr. Bruce D. Trapp for invaluable suggestions. We thank Flow core in Cleveland Clinic Foundation for the flow cytometry experiments. We thank Aishwarya Yenepalli for help with quantification.
This research was supported by grants from the US National Institutes of Health, the Charles A. Dana Foundation, the National Multiple Sclerosis Society, and the Williams Family Fund for MS Research, as well as a Postdoctoral Fellowship from National Multiple Sclerosis Society (to N. Ohno).
The authors have no competing financial interests.
Footnotes
Abbreviations used:
CNS
central nervous system
EAE
experimental autoimmune encephalomyelitis
MDM
monocyte-derived macrophage
MiDM
microglia-derived macrophage
MS
multiple sclerosis
SBF-SEM
serial block-face scanning electron microscopy
References
- Ajami, B., Bennett J.L., Krieger C., Tetzlaff W., and Rossi F.M.. 2007. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10:1538–1543 10.1038/nn2014 [DOI] [PubMed] [Google Scholar]
- Ajami, B., Bennett J.L., Krieger C., McNagny K.M., and Rossi F.M.. 2011. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 14:1142–1149 10.1038/nn.2887 [DOI] [PubMed] [Google Scholar]
- Arif, A., Chatterjee P., Moodt R.A., and Fox P.L.. 2012. Heterotrimeric GAIT complex drives transcript-selective translation inhibition in murine macrophages. Mol. Cell. Biol. 32:5046–5055 10.1128/MCB.01168-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, J., Huitinga I., Zhao W., Lassmann H., Hickey W.F., and Dijkstra C.D.. 1995. The role of macrophages, perivascular cells, and microglial cells in the pathogenesis of experimental autoimmune encephalomyelitis. Glia. 15:437–446 10.1002/glia.440150407 [DOI] [PubMed] [Google Scholar]
- Brück, W., Porada P., Poser S., Rieckmann P., Hanefeld F., Kretzschmar H.A., and Lassmann H.. 1995. Monocyte/macrophage differentiation in early multiple sclerosis lesions. Ann. Neurol. 38:788–796 10.1002/ana.410380514 [DOI] [PubMed] [Google Scholar]
- Butovsky, O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17:131–143 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona, A.E., Pioro E.P., Sasse M.E., Kostenko V., Cardona S.M., Dijkstra I.M., Huang D., Kidd G., Dombrowski S., Dutta R., et al. 2006. Control of microglial neurotoxicity by the fractalkine receptor. Nat. Neurosci. 9:917–924 10.1038/nn1715 [DOI] [PubMed] [Google Scholar]
- Chiu, I.M., Morimoto E.T., Goodarzi H., Liao J.T., O’Keeffe S., Phatnani H.P., Muratet M., Carroll M.C., Levy S., Tavazoie S., et al. 2013. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4:385–401 10.1016/j.celrep.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk, W., and Horstmann H.. 2004. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2:e329 10.1371/journal.pbio.0020329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmazières, A., Sol-Foulon N., and Lubetzki C.. 2012. Changes at the nodal and perinodal axonal domains: a basis for multiple sclerosis pathology? Mult. Scler. 18:133–137 10.1177/1352458511434370 [DOI] [PubMed] [Google Scholar]
- Donnelly, D.J., Longbrake E.E., Shawler T.M., Kigerl K.A., Lai W., Tovar C.A., Ransohoff R.M., and Popovich P.G.. 2011. Deficient CX3CR1 signaling promotes recovery after mouse spinal cord injury by limiting the recruitment and activation of Ly6Clo/iNOS+ macrophages. J. Neurosci. 31:9910–9922 10.1523/JNEUROSCI.2114-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, B., Matyszak M.K., Esiri M.M., and Perry V.H.. 1997. Axonal damage in acute multiple sclerosis lesions. Brain. 120:393–399 10.1093/brain/120.3.393 [DOI] [PubMed] [Google Scholar]
- Fife, B.T., Huffnagle G.B., Kuziel W.A., and Karpus W.J.. 2000. CC chemokine receptor 2 is critical for induction of experimental autoimmune encephalomyelitis. J. Exp. Med. 192:899–905 10.1084/jem.192.6.899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Frederick T.J., Huff T.B., Goings G.E., Miller S.D., and Cheng J.X.. 2011. Paranodal myelin retraction in relapsing experimental autoimmune encephalomyelitis visualized by coherent anti-Stokes Raman scattering microscopy. J. Biomed. Opt. 16:106006 10.1117/1.3638180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. ; Immunological Genome Consortium. 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero, E., Schulz C., and Geissmann F.. 2013. Development and homeostasis of “resident” myeloid cells: the case of the microglia. Glia. 61:112–120 10.1002/glia.22393 [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford, A., and Ravichandran K.S.. 2013. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 5:a008748 10.1101/cshperspect.a008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D., Shi F.D., Jung S., Pien G.C., Wang J., Salazar-Mather T.P., He T.T., Weaver J.T., Ljunggren H.G., Biron C.A., et al. 2006. The neuronal chemokine CX3CL1/fractalkine selectively recruits NK cells that modify experimental autoimmune encephalomyelitis within the central nervous system. FASEB J. 20:896–905 10.1096/fj.05-5465com [DOI] [PubMed] [Google Scholar]
- Huitinga, I., van Rooijen N., de Groot C.J., Uitdehaag B.M., and Dijkstra C.D.. 1990. Suppression of experimental allergic encephalomyelitis in Lewis rats after elimination of macrophages. J. Exp. Med. 172:1025–1033 10.1084/jem.172.4.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitinga, I., Damoiseaux J.G., Döpp E.A., and Dijkstra C.D.. 1993. Treatment with anti-CR3 antibodies ED7 and ED8 suppresses experimental allergic encephalomyelitis in Lewis rats. Eur. J. Immunol. 23:709–715 10.1002/eji.1830230321 [DOI] [PubMed] [Google Scholar]
- Izikson, L., Klein R.S., Charo I.F., Weiner H.L., and Luster A.D.. 2000. Resistance to experimental autoimmune encephalomyelitis in mice lacking the CC chemokine receptor (CCR)2. J. Exp. Med. 192:1075–1080 10.1084/jem.192.7.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisäkk, P., Mahad D.J., Callahan M.K., Sikora K., Trebst C., Tucky B., Wujek J., Ravid R., Staugaitis S.M., Lassmann H., and Ransohoff R.M.. 2004. Expression of CCR7 in multiple sclerosis: implications for CNS immunity. Ann. Neurol. 55:627–638 10.1002/ana.20049 [DOI] [PubMed] [Google Scholar]
- Kulkarni, M.M.2011. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr. Protoc. Mol. Biol. Chapter 25, Unit25B 10 [DOI] [PubMed] [Google Scholar]
- Liu, L., Belkadi A., Darnall L., Hu T., Drescher C., Cotleur A.C., Padovani-Claudio D., He T., Choi K., Lane T.E., et al. 2010. CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: relevance to multiple sclerosis. Nat. Neurosci. 13:319–326 10.1038/nn.2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinl, E., Derfuss T., Krumbholz M., Pröbstel A.K., and Hohlfeld R.. 2011. Humoral autoimmunity in multiple sclerosis. J. Neurol. Sci. 306:180–182 10.1016/j.jns.2010.08.009 [DOI] [PubMed] [Google Scholar]
- Mizutani, M., Pino P.A., Saederup N., Charo I.F., Ransohoff R.M., and Cardona A.E.. 2012. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J. Immunol. 188:29–36 10.4049/jimmunol.1100421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, R., Jia J., Arif A., Ray P.S., and Fox P.L.. 2009. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34:324–331 10.1016/j.tibs.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauta, A.J., Castellano G., Xu W., Woltman A.M., Borrias M.C., Daha M.R., van Kooten C., and Roos A.. 2004. Opsonization with C1q and mannose-binding lectin targets apoptotic cells to dendritic cells. J. Immunol. 173:3044–3050 10.4049/jimmunol.173.5.3044 [DOI] [PubMed] [Google Scholar]
- Perry, V.H., Cunningham C., and Holmes C.. 2007. Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 7:161–167 10.1038/nri2015 [DOI] [PubMed] [Google Scholar]
- Pino, P.A., and Cardona A.E.. 2011. Isolation of brain and spinal cord mononuclear cells using percoll gradients. J. Vis. Exp. 2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff, R.M.2007. Microgliosis: the questions shape the answers. Nat. Neurosci. 10:1507–1509 10.1038/nn1207-1507 [DOI] [PubMed] [Google Scholar]
- Ransohoff, R.M.2012. Animal models of multiple sclerosis: the good, the bad and the bottom line. Nat. Neurosci. 15:1074–1077 10.1038/nn.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup, N., Cardona A.E., Croft K., Mizutani M., Cotleur A.C., Tsou C.L., Ransohoff R.M., and Charo I.F.. 2010. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS ONE. 5:e13693 10.1371/journal.pone.0013693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarin, C., Stohlman S.A., Atkinson R., Ransohoff R.M., and Bergmann C.C.. 2010. Monocytes regulate T cell migration through the glia limitans during acute viral encephalitis. J. Virol. 84:4878–4888 10.1128/JVI.00051-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Serafini, B., Rosicarelli B., Magliozzi R., Stigliano E., Capello E., Mancardi G.L., and Aloisi F.. 2006. Dendritic cells in multiple sclerosis lesions: maturation stage, myelin uptake, and interaction with proliferating T cells. J. Neuropathol. Exp. Neurol. 65:124–141 [DOI] [PubMed] [Google Scholar]
- Steinman, L., Martin R., Bernard C., Conlon P., and Oksenberg J.R.. 2002. Multiple sclerosis: deeper understanding of its pathogenesis reveals new targets for therapy. Annu. Rev. Neurosci. 25:491–505 10.1146/annurev.neuro.25.112701.142913 [DOI] [PubMed] [Google Scholar]
- Stoll, G., Griffin J.W., Li C.Y., and Trapp B.D.. 1989. Wallerian degeneration in the peripheral nervous system: participation of both Schwann cells and macrophages in myelin degradation. J. Neurocytol. 18:671–683 10.1007/BF01187086 [DOI] [PubMed] [Google Scholar]
- Trapp, B.D., Peterson J., Ransohoff R.M., Rudick R., Mörk S., and Bö L.. 1998. Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med. 338:278–285 10.1056/NEJM199801293380502 [DOI] [PubMed] [Google Scholar]
- Trebst, C., Sørensen T.L., Kivisäkk P., Cathcart M.K., Hesselgesser J., Horuk R., Sellebjerg F., Lassmann H., and Ransohoff R.M.. 2001. CCR1+/CCR5+ mononuclear phagocytes accumulate in the central nervous system of patients with multiple sclerosis. Am. J. Pathol. 159:1701–1710 10.1016/S0002-9440(10)63017-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S., Nandi S., Chitu V., Yeung Y.G., Yu W., Huang M., Williams L.T., Lin H., and Stanley E.R.. 2010. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J. Leukoc. Biol. 88:495–505 10.1189/jlb.1209822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, K.C., Ulvestad E., and Hickey W.F.. 1994. Immunology of multiple sclerosis. Clin. Neurosci. 2:229–245 [PubMed] [Google Scholar]
- Wynn, T.A., Chawla A., and Pollard J.W.. 2013. Macrophage biology in development, homeostasis and disease. Nature. 496:445–455 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Li J., Ferguson G.D., Mercurio F., Khambatta G., Morrison L., Lopez-Girona A., Corral L.G., Webb D.R., Bennett B.L., and Xie W.. 2009. Immunomodulatory drugs reorganize cytoskeleton by modulating Rho GTPases. Blood. 114:338–345 10.1182/blood-2009-02-200543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material