Tremor–ataxia with central hypomyelination (TACH) leukodystrophy maps to chromosome 10q22.3–10q23.31 (original) (raw)
. Author manuscript; available in PMC: 2014 Aug 28.
Published in final edited form as: Neurogenetics. 2010 Jul 17;11(4):457–464. doi: 10.1007/s10048-010-0251-8
Abstract
Leukodystrophies are a heterogeneous group of disorders associated with abnormal central nervous system white matter. The clinical features invariably include upper motor neuron signs and developmental regression with or without other neurological manifestations. The objective of this study was to characterize clinically and genetically a new form of childhood-onset leukodystrophy with ataxia and tremor. We recruited seven French-Canadian cases belonging to five families affected by an unknown form of childhood-onset leukodystrophy. Genome-wide scans (GWS) were performed using the Illumina Hap310 or Hap610 Bead Chip to identify regions of shared homozygosity that were further studied for linkage with STS markers. All cases presented between the ages of 1 and 5 years with spasticity along with other upper motor neuron signs, prominent postural tremor, and cerebellar signs. Though motor regression is a constant feature, cognitive functions are relatively preserved, even late in the course of the disease. The higher frequency of founder diseases in the French-Canadian population and the segregation in pedigrees are suggestive of a recessive mode of inheritance. By homozygosity mapping, we established linkage to a 12.6-Mb SNP-haplotyped region on chromosome 10q22.3–10q23.31 (maximum LOD score: 5.47). We describe an autosomal recessive childhood-onset leukodystrophy with ataxia and tremor mapping to a 12.6 Mb interval on chromosome 10q22.3–10q23.31. Identification of the mutated gene will allow precise diagnosis and genetic counseling and shed light on how its perturbed function leads to white matter abnormalities.
Keywords: Leukodystrophy, White matter disease, Hypomyelination, Ataxia, Tremor, Linkage
Introduction
Leukodystrophies are a heterogeneous group of neurodegenerative disorders affecting preferentially the central nervous system white matter. These disorders lead to progressive spasticity and other upper motor neuron signs, as well as developmental and/or cognitive regression [1]. In some instances, patients may have cerebellar ataxia, peripheral nerve involvement, psychiatric symptoms, and movement disorders [1]. A number of leukodystrophies have identified genetic causes with autosomal recessive, autosomal dominant, and X-linked inheritance [1-11]. The French-Canadian population is known for its numerous regional founder effects for recessive diseases [12]. The apparent geographical clustering of recessive French-Canadian cases of an undiagnosed leukodystrophy with ataxic features led to a search for a potential new chromosomal locus by homozygosity mapping. We report the clinical features of a new form of childhood-onset leukodystrophy and its mapping to chromosome 10q22.3–10q23.31.
Subjects and methods
Clinical evaluation and DNA isolation
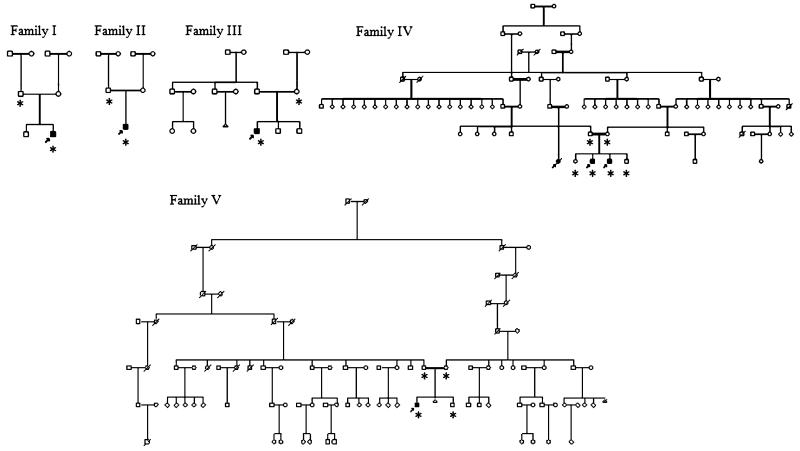
We have identified a cohort of six living and one deceased cases, from five different French-Canadian families (Fig. 1), presenting in early childhood with progressive motor deterioration, spasticity, tremor, and gait ataxia. All living cases underwent a detailed neurological examination by experienced neurologists. A detailed chart review was performed for all cases, including case 6. This patient died at age 21; her parents refused the autopsy. Neuroimaging was reviewed for all cases by A.V., although for cases 3 and 6, a restricted number of images were available. This project was approved by the institutional Ethics Committee of the Centre de Recherche du CHUM (CRCHUM). Informed consent was obtained from all participants or parents. Genomic DNA was extracted from peripheral blood lymphocytes using a standard method or from saliva using the Oragene DNA extraction kit (DNA Genotek, Ottawa, Canada).
Fig. 1.
Pedigrees of the five French-Canadian families with tremor–ataxia with central hypomyelination (TACH), including consanguineous families 4 and 5. Affected cases are shaded black and participants for which a genome scan was performed are marked with an asterisk
Genome scan and linkage analysis
Genome-wide scans using the Illumina Hap310 or Hap610 Bead chip (310 or 610 K SNPs, respectively) were conducted at the McGill University and Genome Quebec Innovation Centre (Montreal, Canada) on all living cases and at least one non-affected family member per family. Multipoint linkage analysis was performed using GENE-HUNTER v.2.1 and Simwalk2 v.2.91 [13]. The files used for Simwalk2 were produced with Mega2 v.4.3 [14]. Marker order and genetic distances were based on the deCODE genetic map and UCSC physical map (http://genome.ucsc.edu, March 2006 assembly). For the linkage analyses, allele frequencies were calculated from 162 control chromosomes of non-affected French-Canadian from the Quebec province. The leukodystrophy phenotype was analyzed as an autosomal recessive trait with 100% penetrance, with an estimate disease gene frequency of 0.001 and no phenocopy.
Exclusion of candidate genes
We sequenced the coding sequence, 5′ and 3′ UTR of the following ten genes in the 12.6-Mb candidate interval: PPIF (peptidylprolyl isomerase F precursor), ANXA11 (annexin A11), ATAD1 (ATPase family, AAA domain containing 1), SFTPD (pulmonary surfactant-associated protein D), CH25H (cholesterol 25-hydroxylase), EIF5AL1 (eukaryotic translation initiation factor 5A-like), SFTPA1 (surfactant protein A1 precursor), SFTPA2 (surfactant protein A2 precursor), LRRC21 (leucine-rich repeat containing protein 21 precursor), and NRG3 (neuregulin 3). Candidate genes were selected for sequencing based on their expression profile (www.cgl.ucsf.edu/cgi-bin/genentech/genehub-gepis/), gene ontology (http://genome.ucsc.edu/), and using the SUSPECTS program (www.genetics.med.ed.ac.uk/suspects/). PCR primers (supplementary Table 1) were designed using ExonPrimer (http://genome.ucsc.edu) or Primer3 (http//frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). PCR reactions and sequencing reactions were conducted at the McGill University and Genome Quebec Innovation Centre (Montreal, Canada). Sequences were analyzed using SeqMan 4.03 (DNAStar, Madison, WI, USA), Chromas 1.62 (Technelysium, Helensvale, Australia), and Mutation Surveyor v.3.1 (SoftGenetics, State College, PA, USA).
Results
Clinical features
We recruited a cohort of 14 French-Canadian cases of leukodystrophy of unknown cause. Seven cases belonging to five families were considered to be likely affected by the same form of leukodystrophy based on their clinical and radiological characteristics. The sex ratio was six boys for one girl (Fig. 1). In family 4, the presence of a deceased distant affected female cousin (case 6) of two brothers (cases 4 and 5) favors a recessive rather than an X-linked mode of inheritance. Further support for a recessive mode of transmission comes from the observation that, in families 4 and 5, the parents are known to be distantly related and both originate from the contiguous Bellechasse and Beauce regions where another recessive founder effect has been described for a recessive ataxia (Fig. 1) [15]. The mother of case 2 also originates from the Beauce region. This micro-regional cluster further raised the possibility of a recessive founder mutation shared by these three families. Families 1 and 3 originate from the Gaspésie region also known for its regional founder effects [16,17]. The clinical features of the seven cases are summarized in Table 1. The average age of onset is 2.5 years (1–5 years), although age of onset was variable even within a single family (family 4). All cases presented with motor regression, ataxia, and tremor (Table 1). Gait was clearly modified by a combination of pyramidal spasticity and cerebellar ataxia. The gait ataxia is associated with a prominent tremor of the upper extremities, which is present both with posture holding and action, dysarthria, and abnormal ocular saccades and/or pursuits with or without gaze-evoked nystagmus. As the disease progresses, the patients’ motor difficulties worsened and lead to the use of a wheelchair between ages 4 and 12 years (average 7.3 years). The exception is case 5, with a later disease onset at age 5, who is still ambulant and walking without any aid at age 12. Additional features include a history of deterioration with infections or stress (3/7, 43%), bulbar symptoms such as dysphagia (4/7, 57%), and drooling (5/7, 71%), optic atrophy (4/7, 57%), and partial complex seizures (2/7, 29%). Cognitive decline is mild and occurs later in the course, but was clearly observed in 6/7 cases. Of interest, two out of seven cases have some degree of hypodontia; case 4 is missing four teeth (teeth #12–22–31–41) and case 5 is missing one (tooth #22). One case (#7) has documented hypogonadotropic hypogonadism but no hypodontia.
Table 1.
Clinical features
| Patient # | Family # | Gender | Developmentaldelay | Age ofonset (y) | Age in 2010 | Ataxia | Dysmetria | Spasticity | Hyperreflexia | Babinski | Dysarthria | Dysphagia |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | M | + | 1 | 6 | + | + | + | + | + | + | |
| 2 | 2 | M | 2.5 | 14 | + | + | + | + | + | + | + | |
| 3 | 3 | M | + | 2 | 10 | + | + | + | + | + | + | |
| 4 | 4 | M | + | 1 | 15 | + | + | + | + | + | + | + |
| 5 | 4 | M | + | 5 | 13 | + | + | + | + | + | ||
| 6 | 4 | F | 3 | N/A | + | + | + | + | + | + | + | |
| 7 | 5 | M | 3 | 28 | + | + | + | + | + | + | + | |
| Mean | 2.5 |
| Patient # | Drooling | Tremor | Gaze-evokednystagmus | Opticatrophy | Bladderdysfunction | Seizures | Wheelchair | Age—wheelchair (y) | Neuropathy | Cognitiveregression (mild) | Hypodontia | Hypogonadotropichypogonadism | Age ofdeath |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | + | + | N/A | + | + | 4 | ||||||
| 2 | + | + | + | + | + | + | + | 7 | N/A | + | |||
| 3 | + | + | + | + | 5 | + | |||||||
| 4 | + | + | + | + | + | + | + | 8 | + | + | |||
| 5 | + | + | + | ||||||||||
| 6 | + | + | + | + | + | 8 | + | 21 | |||||
| 7 | + | + | + | + | + | + | 12 | + | + | ||||
| Mean | 7.3 |
All cases in the cohort have had extensive workups in different institutions. The following leukodystrophies were excluded in all living patients: metachromatic leukodystrophy, Krabbe, Canavan, and adrenoleukodystrophy. Nerve conduction studies and electromyograms were performed in all seven patients and did not reveal any abnormalities. Sural nerve biopsies on cases 6 and 7 were normal, including electronic microscopy for case 7. Mutation screening was performed on the following genes in at least one individual: GJA12 (case 3), EIF2B1_–_EIF2B2_–_EIF2B3_–_EIF2B4_–_EIF2B5 (cases 3 and 4), PLP1 (case 7), NPC1 and NPC2 (cases 3 and 7), and GFAP (cases 3 and 4). Though the tremor–ataxia with central hypomyelination (TACH) locus lies 5.1 Mb telomeric to the PSAP gene, we decided to exclude it by sequencing. This gene encodes the four saposins: A, B, C, and D. Mutations of the PSAP gene can lead to deficiency in all saposins (combined saposin deficiency), in saposin A, B, C, or D, leading, respectively, to atypical Krabbe, metachromatic leukodystrophy variant, atypical Gaucher, and Tay-Sachs variant [18-22]. No mutation in this gene was found in case 4 and the urine sulfatides were normal in cases 3 and 4, excluding this gene as responsible for TACH. FISH study for PLP1 duplication was performed in two cases (cases 3 and 4). Sialic acid was measured in four cases (cases 1, 4, 5, and 6). These extensive investigations failed to uncover the cause for the leukodystrophy in all seven participants. The only frequent abnormality shared by six of the seven participants was the presence of high total cholesterol on the lipid profile, from a mild augmentation to an elevation significant enough to require treatment with HMG-CoA reductase inhibitor in case 7.
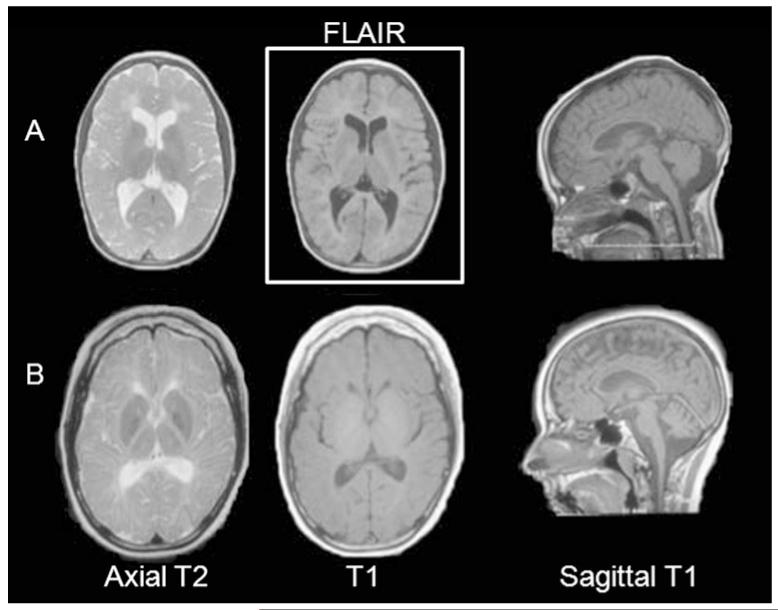
Available neuroimaging was reviewed for all seven cases, although only a small number of sequences were available for cases 3 and 6. The most typical MRI findings of the least (case 5) and most clinically affected (case 7) patients are illustrated in Fig. 3. All cases have a hypomyelinating leukoencephalopathy [23], with hyper or isointense appearing white matter on T1 and concomitant hyperintense white matter structures on T2. White matter abnormalities are diffuse in nature but with relative sparing of arcuate fibers and optic radiations in the less affected cases, involving the periventricular and deep white matter preferentially. The patients were also found to have increased cerebrospinal fluid spaces within the posterior fossa, as well as thinning of the corpus callosum. In the most severely clinically affected patients, there is minimal rarefaction of the deep white matter near the frontal and occipital ventricular horns on FLAIR imaging, as well as cerebellar atrophy.
Fig. 3.
MRI features in tremor–ataxia with central hypomyelination (TACH). Characteristic MRI images in two subjects: cases 5 (a) and 7 (b) (the least and most affected patients, at ages 11 and 21 years, respectively). The MRI show diffuse hypomyelination with diffusely increased T2 signal, isointense or hyperintense T1 signal, with relative sparing of arcuate fibers and optic radiations. Thinning of the corpus callosum is a constant feature. In the most clinically affected patients, such as in case 7, there is some periventricular deep white matter rarefaction on FLAIR images, as well as atrophy of the cerebellum
Mapping of the locus and sequencing of candidate genes
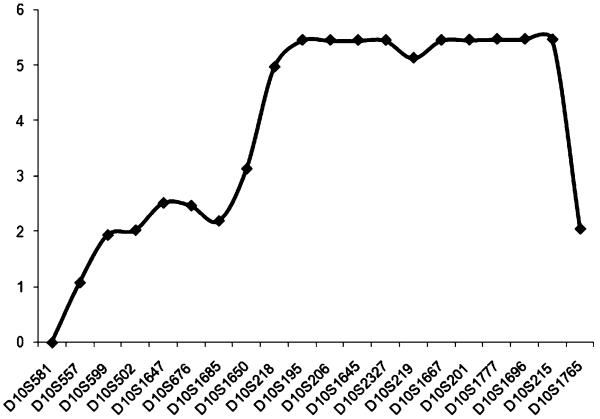
DNA samples from the six living cases, at least one unaffected parent, and in cases 4, 5, and 7, siblings, were sent to McGill University and Genome Quebec Innovation Centre (Montreal, Canada) for a genome-wide scan (GWS) using the Illumina Hap310 or Hap610 Bead chip (310 or 610 K SNPs). The affected children of families 4 and 5 were found to be homozygotes for 12.6 Mb (3,000 SNPs) on chromosome 10q22.3–q23.31. Further genotyping with microsatellite (STS) markers was performed in this region. The three cases (4, 5, and 7) were found to be homozygous for all markers in this region. A multipoint cumulative LOD score of 5.47 was obtained for STS markers D10S201, D10S1777, D10S1696, and using Simwalk2 v.2.91 and Gene Hunter v.2.1 for families 4 and 5 (Fig. 2). Cases 1, 2, and 3 from families 1, 2, and 3 were found to be compound heterozygotes for the shared haplotype of families 4 and 5. This finding suggests that at least one other mutation is present in our group of patients. These results are compatible with the geographical origins of these three families (1, 2, and 3), the parents being unrelated and from different regions in Quebec. This 12.6-Mb (rs7069982–rs2071510) locus on 10q22.3–q23.31 has not been previously associated with a leukodystrophy.
Fig. 2.
Multipoint LOD scores generated by the analysis of family 4 and family 5 for 20 markers on chromosome 10q21.2–10q23.2 markers
Sixty-one genes lie in the 12.6-Mb candidate interval. Investigation of changes in gene copy number using the SNP genotype generated during the GWS was performed using copy number variation (CNV) analysis with BeadStudio v3.1 (Applied Biosystems, Foster City, CA, USA) and using a propriety program of six algorithms for high-throughput detection of CNV. No putative pathogenic CNV were detected in the candidate interval. Sequencing analysis of all coding and surrounding intronic sequences, 5′ and 3′ UTRs were performed for the following ten candidate genes lying in the 12.6-Mb region: PPIF, ANXA11, ATAD1, SFTPD, CH25H, EIF5AL1, SFTPA1, SFTPA2, LRRC21, and NRG3. The genes were selected because their expression profiles were similar to known causal leukodystrophy genes and/or because of their presumed interaction(s) with these genes (http://www.genetics.med.ed.ac.uk/suspects/). The EIF5AL1 gene was selected as one of our prime candidates because it is part of the elongation factor family of genes as the following mutated genes in CACH: EIF2B1, EIF2B2, EIF2B3, EIF2B4, and EIF2B5 [1]. PPIF and SFTPD are respectively predicted to have a similar expression profile and to have a shared function with genes belonging to the elongation factors family. The genes SFTPD, SFTPA1, SFTPA2, and ANXA11 are predicted to share different functions with the PSAP gene (prosaposin isoform A preproprotein), a gene known to cause rare forms of Krabbe and metachromatic leukodystrophies [5]. The SFTPD gene is also presumed to share some of the lysosomal functions of the ARSA (arylsulfatase A) gene. The CH25H gene was chosen based on its role in cholesterol metabolism, considering that six of the seven cases have elevated total cholesterol. Finally, the LRRC21 and NRG3 genes were selected because of their expression profiles in the brain. No mutations were uncovered in any of these genes.
Conclusion
We describe a new form of recessive childhood-onset leukodystrophy, TACH, and map its locus to chromosome 10q22.3–10q23.31. This leukodystrophy presents around the same ages as the typical forms of metachromatic leukodystrophy and CACH (18 to 24 months and 12 months to 5 years of age, respectively) [1]. The clinical features are somewhat similar to other forms of leukodystrophies presenting in early childhood, with progressive motor deterioration, spasticity, and other upper motor neuron signs. The more typical clinical characteristics of these cases are the presence of prominent ataxia and of an important postural and action tremor. To underline this clinical observation, we chose to call it TACH. We believe that this disease is better characterized as a leukodystrophy than an ataxia because of the universal white matter abnormalities on MRI studies (Fig. 3) and because the clinical presentation is more complex, involving several other neurological systems. Its clinical presentation resembles CACH in several ways, with ataxia being an important clinical finding [1]; however, MRI features are distinct from this disorder [24]. The MRI findings of TACH are more consistent with the group of disorders characterized by hypomyelination, including those caused by GJA12/GJC2 mutations or Pelizaeus-Merzbacher-like disease [11], HCC, or hypomyelination with congenital cataracts [25], and 4H or hypomyelination with hypodontia and hypogonadotropic hypogonadism, among others. Pelizaeus-Merzbacher-like disease was ruled out by sequencing the GJA12 gene. HCC was eliminated as a possible diagnosis in our cases because of inconsistent clinical features and different loci. The fact that the oldest case is treated for hypogonadotropic hypogonadism and that two participants are known for hypodontia raised the possibility that these patients may have a variant of 4H syndrome: hypomyelination, hypogonadotropic hypogonadism, and hypodontia [26-28]. The 4H syndrome was first described in 2005 [29] in four unrelated girls with progressive ataxia, hypomyelination, and delayed dentition. Since the first description, a few more patients have been reported [27-30]. It is thought to be inherited in an autosomal recessive fashion, but the chromosomal locus is still unknown. Though there appears to be some clinical overlap between 4H and TACH, none of our cases have the full set of features. Only two out seven have missing teeth, and a single patient has hypogonadotropic hypogonadism. The two sural nerve biopsies on cases 6 and 7 were interpreted as normal. The nerve biopsy of case 7 was studied by electron microscopy and no abnormality supportive of a 4H diagnosis was uncovered. Nerve biopsies of patients with the 4H syndrome were reported to demonstrate clefts lined with granular debris, expanded abaxonal space, outpocketing with vacuolar disruption, and loss of normal myelin periodicity [27]. The identification of the 4H locus or the gene for TACH will establish if the two disorders are allelic. To identify the mutated gene responsible for TACH, further genomic analyses will be required to further fine map the candidate region by recruiting additional families from Quebec and other countries. The identification of the TACH gene will contribute to our understanding of the mechanisms leading to inherited white matter diseases. It will also allow clinicians to offer definite diagnoses for one form of the growing number of leukodystrophies and genetic counseling to their families.
Supplementary Material
Vanderver_SD
Acknowledgements
We wish to thank all family members for their participation. We would also like to thank all the clinicians who referred patients to us and who were not included in this article: Drs. Marie-Emmanuelle Dilenge, Chantal Poulin, Michael Shevell, Amelie Nadeau, Bruno Maranda, Renée-Myriam Boucher, and Jean Mathieu. We also wish to thank Alexandre Montpetit and Pierre Lepage from McGill University and Genome Quebec Innovation Center for their technical expertise. Dr. Bernard has received a scholarship grant from the Réseau de medicine génique appliquée (RMGA) and from the Fonds de Recherche en Santé du Québec (FRSQ). I. Thiffault has received a scholarship grant from CIHR and ETP fellowship from the National Bank Financial Group. Dr. Vanderver has received funding from the American Academy of Neurology Foundation Clinical Research Training Fellowship program.
Funding information This research project was supported by the Quebec-base foundation: “_Fondation sur les leukodystrophies_” (http://www.leucofondation.com/).
Footnotes
Integrity of research and reporting Ethical standards The experiments comply with the current laws of the country in which they were performed.
Conflict of interest The authors declare that they have no conflicts of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s10048-010-0251-8) contains supplementary material, which is available to authorized users.
Contributor Information
Geneviève Bernard, Laboratoire de neurogénétique de la motricité, Neuromics Center for Excellence of Université de Montréal, CRCHUM, 1560 Sherbrooke East, Montreal, Quebec H2L 4M1, Canada.
Isabelle Thiffault, Laboratoire de neurogénétique de la motricité, Neuromics Center for Excellence of Université de Montréal, CRCHUM, 1560 Sherbrooke East, Montreal, Quebec H2L 4M1, Canada.
Martine Tetreault, Laboratoire de neurogénétique de la motricité, Neuromics Center for Excellence of Université de Montréal, CRCHUM, 1560 Sherbrooke East, Montreal, Quebec H2L 4M1, Canada.
Maria Lisa Putorti, Laboratoire de neurogénétique de la motricité, Neuromics Center for Excellence of Université de Montréal, CRCHUM, 1560 Sherbrooke East, Montreal, Quebec H2L 4M1, Canada.
Isabelle Bouchard, CHUQ, Centre Mère-Enfant, 2705 Boul. Laurier, Quebec, Quebec G1R 2J6, Canada.
Michel Sylvain, CHUQ, Centre Mère-Enfant, 2705 Boul. Laurier, Quebec, Quebec G1R 2J6, Canada.
Serge Melançon, Montreal Children’s Hospital, McGill University, 2300 Tupper, Montreal, Quebec H3H 1P3, Canada.
Rachel Laframboise, CHUQ, Centre Mère-Enfant, 2705 Boul. Laurier, Quebec, Quebec G1R 2J6, Canada.
Pierre Langevin, CHUQ, Centre Mère-Enfant, 2705 Boul. Laurier, Quebec, Quebec G1R 2J6, Canada.
Jean-Pierre Bouchard, Department of Neurological Sciences, CHA-Hôpital Enfant-Jésus, 1401, 18e rue, Quebec, Quebec G1J 1Z4, Canada.
Michel Vanasse, Neurology Department, CHU-Hôpital Ste-Justine, 3175, Chemin de la Côte-Sainte-Catherine, Montreal, Quebec H3T 1C5, Canada.
Adeline Vanderver, Children’s National Medical Center, Children’s Research Institute, Center for Genetic Medicine Research, 111 Michigan Ave, Washington DC 20010, USA.
Guillaume Sébire, CHUS, Sherbrooke University, 3001, 12e Avenue Nord, Sherbrooke, Quebec J1H 5N4, Canada.
Bernard Brais, Laboratoire de neurogénétique de la motricité, Neuromics Center for Excellence of Université de Montréal, CRCHUM, 1560 Sherbrooke East, Montreal, Quebec H2L 4M1, Canada; Laboratoire de neurogénétique de la motricité, M4211-L3, Hôpital Notre-Dame-CHUM, CRCHUM, 1560 Sherbrooke East, Montreal, H2L 4M1, Quebec, Canada.
References
- 1.Schiffmann R, van der Knaap MS. The latest on leukodystrophies. Curr Opin Neurol. 2004;17:187–192. doi: 10.1097/00019052-200404000-00017. [DOI] [PubMed] [Google Scholar]
- 2.Brenner M, Johnson AB, Boespflug-Tanguy O, Rodriguez D, Goldman JE, Messing A. Mutations in GFAP, encoding glial fibrillary acidic protein, are associated with Alexander disease. Nat Genet. 2001;27:117–120. doi: 10.1038/83679. [DOI] [PubMed] [Google Scholar]
- 3.Cannizzaro LA, Chen YQ, Rafi MA, Wenger DA. Regional mapping of the human galactocerebrosidase gene (GALC) to 14q31 by in situ hybridization. Cytogenet Cell Genet. 1994;66:244–245. doi: 10.1159/000133703. [DOI] [PubMed] [Google Scholar]
- 4.Gieselmann V, Polten A, Kreysing J, von Figura K. Molecular genetics of metachromatic leukodystrophy. J Inherit Metab Dis. 1994;17:500–509. doi: 10.1007/BF00711364. [DOI] [PubMed] [Google Scholar]
- 5.Henseler M, Klein A, Reber M, Vanier MT, Landrieu P, Sandhoff K. Analysis of a splice-site mutation in the sap-precursor gene of a patient with metachromatic leukodystrophy. Am J Hum Genet. 1996;58:65–74. [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6:1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- 7.Kaul R, Gao GP, Balamurugan K, Matalon R. Cloning of the human aspartoacylase cDNA and a common missense mutation in Canavan disease. Nat Genet. 1993;5:118–123. doi: 10.1038/ng1093-118. [DOI] [PubMed] [Google Scholar]
- 8.Leegwater PA, Yuan BQ, van der Steen J, Mulders J, Konst AA, Boor PK, Mejaski-Bosnjak V, van der Maarel SM, Frants RR, Oudejans CB, Schutgens RB, Pronk JC, van der Knaap MS. Mutations of MLC1 (KIAA0027), encoding a putative membrane protein, cause megalencephalic leukoencephalopathy with subcortical cysts. Am J Hum Genet. 2001;68:831–838. doi: 10.1086/319519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosser J, Douar AM, Sarde CO, Kioschis P, Feil R, Moser H, Poustka AM, Mandel JL, Aubourg P. Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature. 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- 10.Orcesi S, La PR, Fazzi E. Aicardi-Goutieres syndrome. Br Med Bull. 2009;89:183–201. doi: 10.1093/bmb/ldn049. [DOI] [PubMed] [Google Scholar]
- 11.Uhlenberg B, Schuelke M, Ruschendorf F, Ruf N, Kaindl AM, Henneke M, Thiele H, Stoltenburg-Didinger G, Aksu F, Topaloglu H, Nurnberg P, Hubner C, Weschke B, Gartner J. Mutations in the gene encoding gap junction protein alpha 12 (connexin 46.6) cause Pelizaeus-Merzbacher-like disease. Am J Hum Genet. 2004;75:251–260. doi: 10.1086/422763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laberge AM, Michaud J, Richter A, Lemyre E, Lambert M, Brais B, Mitchell GA. Population history and its impact on medical genetics in Quebec. Clin Genet. 2005;68:287–301. doi: 10.1111/j.1399-0004.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 13.Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52:121–131. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- 14.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 15.Dupre N, Gros-Louis F, Chrestian N, Verreault S, Brunet D, de Verteuil D, Brais B, Bouchard JP, Rouleau GA. Clinical and genetic study of autosomal recessive cerebellar ataxia type 1. Ann Neurol. 2007;62:93–98. doi: 10.1002/ana.21143. [DOI] [PubMed] [Google Scholar]
- 16.Duquette A, Roddier K, McNabb-Baltar J, Gosselin I, St-Denis A, Dicaire MJ, Loisel L, Labuda D, Marchand L, Mathieu J, Bouchard JP, Brais B. Mutations in senataxin responsible for Quebec cluster of ataxia with neuropathy. Ann Neurol. 2005;57:408–414. doi: 10.1002/ana.20408. [DOI] [PubMed] [Google Scholar]
- 17.Gosselin I, Thiffault I, Tetreault M, Chau V, Dicaire MJ, Loisel L, Emond M, Senderek J, Mathieu J, Dupre N, Vanasse M, Puymirat J, Brais B. Founder SH3TC2 mutations are responsible for a CMT4C French-Canadians cluster. Neuromuscul Disord. 2008;18:483–492. doi: 10.1016/j.nmd.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Christomanou H, Chabas A, Pampols T, Guardiola A. Activator protein deficient Gaucher’s disease. A second patient with the newly identified lipid storage disorder. Klin Wochenschr. 1989;67:999–1003. doi: 10.1007/BF01716064. [DOI] [PubMed] [Google Scholar]
- 19.Harzer K, Paton BC, Poulos A, Kustermann-Kuhn B, Roggendorf W, Grisar T, Popp M. Sphingolipid activator protein deficiency in a 16-week-old atypical Gaucher disease patient and his fetal sibling: biochemical signs of combined sphingolipidoses. Eur J Pediatr. 1989;149:31–39. doi: 10.1007/BF02024331. [DOI] [PubMed] [Google Scholar]
- 20.Kretz KA, Carson GS, Morimoto S, Kishimoto Y, Fluharty AL, O’Brien JS. Characterization of a mutation in a family with saposin B deficiency: a glycosylation site defect. Proc Natl Acad Sci USA. 1990;87:2541–2544. doi: 10.1073/pnas.87.7.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnabel D, Schroder M, Furst W, Klein A, Hurwitz R, Zenk T, Weber J, Harzer K, Paton BC, Poulos A. Simultaneous deficiency of sphingolipid activator proteins 1 and 2 is caused by a mutation in the initiation codon of their common gene. J Biol Chem. 1992;267:3312–3315. [PubMed] [Google Scholar]
- 22.Spiegel R, Bach G, Sury V, Mengistu G, Meidan B, Shalev S, Shneor Y, Mandel H, Zeigler M. A mutation in the saposin A coding region of the prosaposin gene in an infant presenting as Krabbe disease: first report of saposin A deficiency in humans. Mol Genet Metab. 2005;84:160–166. doi: 10.1016/j.ymgme.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Schiffmann R, van der Knaap MS. Invited article: an MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72:750–759. doi: 10.1212/01.wnl.0000343049.00540.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Knaap MS, Pronk JC, Scheper GC. Vanishing white matter disease. Lancet Neurol. 2006;5:413–423. doi: 10.1016/S1474-4422(06)70440-9. [DOI] [PubMed] [Google Scholar]
- 25.Zara F, Biancheri R, Bruno C, Bordo L, Assereto S, Gazzerro E, Sotgia F, Wang XB, Gianotti S, Stringara S, Pedemonte M, Uziel G, Rossi A, Schenone A, Tortori-Donati P, van der Knaap MS, Lisanti MP, Minetti C. Deficiency of hyccin, a newly identified membrane protein, causes hypomyelination and congenital cataract. Nat Genet. 2006;38:1111–1113. doi: 10.1038/ng1870. [DOI] [PubMed] [Google Scholar]
- 26.Bekiesinska-Figatowska M, Mierzewska H, Kuczynska-Zardzewialy A, Szczepanik E, Obersztyn E. Hypomyelination, hypogonadotropic hypogonadism, hypodontia—First Polish patient. Brain Dev. 2010;32(7):574–578. doi: 10.1016/j.braindev.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Timmons M, Tsokos M, Asab MA, Seminara SB, Zirzow GC, Kaneski CR, Heiss JD, van der Knaap MS, Vanier MT, Schiffmann R, Wong K. Peripheral and central hypomyelination with hypogonadotropic hypogonadism and hypodontia. Neurology. 2006;67:2066–2069. doi: 10.1212/01.wnl.0000247666.28904.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez-Lopez M, Ruiz-Martin Y, de Castro-Castro P, Garzo-Fernandez C, Martin-del VF, Marquez-de la Plata L. Central hypomyelination, hypogonadotrophic hypogonadism and hypodontia: a new leukodystrophy. Rev Neurol. 2008;47:204–208. [PubMed] [Google Scholar]
- 29.Wolf NI, Harting I, Boltshauser E, Wiegand G, Koch MJ, Schmitt-Mechelke T, Martin E, Zschocke J, Uhlenberg B, Hoffmann GF, Weber L, Ebinger F, Rating D. Leukoencephalopathy with ataxia, hypodontia, and hypomyelination. Neurology. 2005;64:1461–1464. doi: 10.1212/01.WNL.0000158615.56071.E3. [DOI] [PubMed] [Google Scholar]
- 30.Wolf NI, Harting I, Innes AM, Patzer S, Zeitler P, Schneider A, Wolff A, Baier K, Zschocke J, Ebinger F, Boltshauser E, Rating D. Ataxia, delayed dentition and hypomyelination: a novel leukoencephalopathy. Neuropediatrics. 2007;38:64–70. doi: 10.1055/s-2007-985137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Vanderver_SD