Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System (original) (raw)
Abstract
The CRISPR/Cas9 system is highly efficient at generating targeted mutations in stable transgenic tomato plants, and homozygous deletions of a desired size can be created in the first generation.
During the past 12 years, there has been rapid development of genome-editing strategies that make it possible to directly target regions of genes in a DNA sequence-specific manner. Two of these strategies, zinc finger nucleases (Urnov et al., 2010) and transcription activator-like nucleases (TALENs; Joung and Sander, 2013), are based on protein-DNA interactions, whereas a third technology, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated9 (Cas9) endonuclease (Liu and Fan, 2014), is an RNA-guided DNA endonuclease system. Each technology has advantages and disadvantages with regard to cost, ease of construction, efficiency of targeting, and specificity (Chen and Gao, 2014). Of the three genome-editing approaches, CRISPR/Cas9 has seen a meteoric rise during the past 2 years, with applications to bacterial, animal, human, and, most recently, plant systems (Pennisi, 2013). This is due, in part, to a greater number of advantages of CRISPR/Cas9, including the straightforward construct design and assembly, as compared with zinc finger nucleases and TALENs.
The first reports of CRISPR/Cas9 editing in plants appeared in 2013, with successful application for both transient expression and recovery of stable transgenic lines. In addition to demonstration of the efficacy in the models Arabidopsis (Arabidopsis thaliana; Li et al., 2013) and Nicotiana benthamiana (Nekrasov et al., 2013), there have also been reports for three crop species, rice (Oryza sativa; Zhang et al., 2014), sorghum (Sorghum bicolor; Jiang et al., 2013), and wheat (Triticum aestivum; Wang et al., 2014). For stable transgenic lines, modifications reported for genes in primary transformants of Arabidopsis and rice were shown to persist into the next generation (Feng et al., 2014; Zhang et al., 2014). Thus, CRISPR/Cas9 is rapidly becoming the tool of choice for gene editing in plants, but further testing is needed to determine whether efficacy will be universal.
Tomato (Solanum lycopersicum) is an ideal candidate for testing CRISPR/Cas9 gene editing in a dicot crop, because of the availability of efficient transformation methodology (Van Eck et al., 2006), the diploidy of the genome, a high-quality genome sequence (Tomato Genome Consortium, 2012), and its economic importance (fresh and processed), which in the United States alone accounts for more than $2 billion (http://www.ers.usda.gov/topics/crops/vegetables-pulses/tomatoes.aspx#.U85tkoBdVSY).
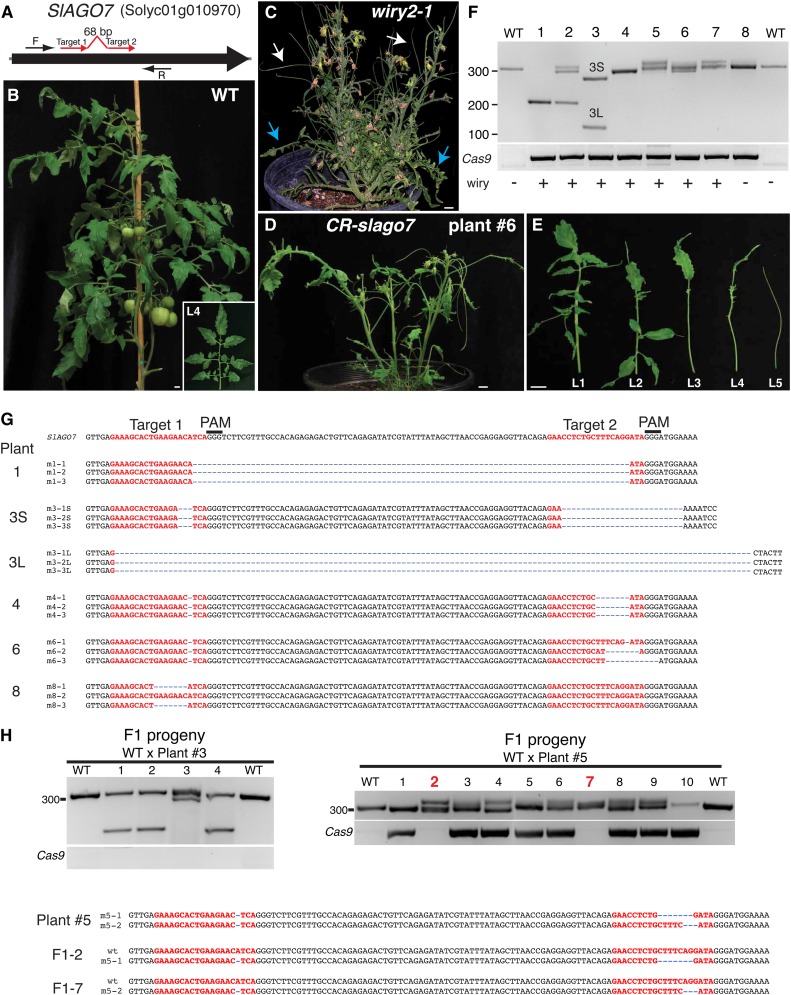
To test the efficacy of CRISPR/Cas9 in tomato, we chose to target a gene that, when function was disrupted, would result in a distinctive, immediately recognizable phenotype early in the plant tissue culture phase of _Agrobacterium tumefaciens_-mediated transformation. A CRISPR/Cas9 construct was designed to target neighboring sequences in the second exon of the tomato homolog of Arabidopsis ARGONAUTE7 (SlAGO7), because loss-of-function mutations are recessive and result in plants whose typical compound flat leaves become needle like or wiry (Fig. 1; Lesley, 1928; Yifhar et al., 2012). SlAGO7 is required for the biogenesis of a class of small RNAs known as transacting short interfering RNAs, which regulate organ polarity through posttranscriptional silencing of AUXIN RESPONSE FACTOR (ARF) genes (Husbands et al., 2009). Strong alleles of slago7 thus produce lower levels of transacting short interfering RNAs and reduced ARF mRNA degradation, resulting in the first leaves of mutant plants having leaflets without petioles and later-formed leaves lacking laminae (Fig. 1C). These distinctive phenotypes allowed us to immediately identify first-generation transformed (T0) plants in which both alleles of SlAGO7 might be mutated.
Figure 1.
CRISPR/Cas9-mediated gene editing in stable transgenic tomato plants. A, Schematic illustrating two sgRNAs (red arrows) targeting the SlAGO7 coding sequence. Cas9/sgRNA1/sgRNA2 were expressed from the same plasmid (Belhaj et al., 2013). Black arrows indicate PCR primers used to evaluate mutation type and efficiency. B, A wild-type (WT) tomato plant at 9 weeks of age, and the fourth-produced leaf from the primary shoot (inset). C, The strong wiry2-1 allele of slago7. First formed leaves have leaflets lacking petioles (blue arrows), and later formed leaves are radialized (white arrows). D and E, A representative CRISPR/Cas9-slago7 (CR-slago7) plant (D) and its first five leaves showing the distinctive loss-of-function recessive wiry syndrome (E). Bars = 1 cm. F and G, PCR genotyping of eight representative CR-slago7 plants showing the type of DNA lesions generated, including homozygosity for the expected 90-bp deletion (plant 1). PCR for the presence of Cas9 is also shown along with wiry phenotypic evaluations. A range of indels was found in the other T0 plants. The PCR products from a subset of plants were cloned and sequenced to validate the deletion in plant 1 and to characterize additional DNA lesions (G). Note that the two deletions in plant 3 are smaller (3S) and larger (3L) than expected. H, Germline transmission and heritability of the small and large deletions from plant 3 (left) and two small deletions from plant 5 (right) in the absence of the inductive Cas9/sgRNA1/sgRNA2 transgene. All four progeny from plant 3 lacked the CRISPR/Cas9 transgene, and all four were heterozygous for deletions, three of which were heterozygous for the large deletion and one heterozygous for the smaller deletion. Of the 10 progeny from plant 5, two plants lacked the CRISPR/Cas9 transgene, and sequencing showed that both were heterozygous for the wild-type allele and one of the two deletions (bottom).
The CRISPR/Cas9 construct we designed to target SlAGO7 contained two single guide RNAs (sgRNAs) with the intention to create large, defined deletions (Belhaj et al., 2013). In addition, we chose target regions of the SlAGO7 sequence that contained the requisite binding region, or protospacer adjacent motif (PAM), for Cas9 cleavage, which is a 5′-NGG located immediately after the 20-bp target DNA. We generated 29 T0 plants and confirmed by PCR that each carried an integrated transfer DNA (T-DNA) from the introduced CRISPR/Cas9 construct. Remarkably, 14 plants (48%) were indistinguishable from a known strong loss-of-function ethyl methanesulfonate-derived slago7 allele (wiry2-1; Yifhar et al., 2012), suggesting a high efficiency of CRISPR/Cas9-mediated targeted mutagenesis in SlAGO7 (Fig. 1, B–E).
To evaluate the types of mutations generated, we performed PCR on eight representative T0 plants using primers that flanked both sgRNA targets (Fig. 1A, black arrows). One plant was homozygous for the expected 90-bp deletion, and as predicted for Cas/sgRNA DNA cleavages (Jinek et al., 2012), cloning and sequencing showed that the deletion breakpoints were located 3 bp upstream of the PAM sites placed right after each of the two sgRNA targets (Fig. 1, F and G). Two additional plants were chimeric, having an allele with the expected deletion and other alleles with smaller deletions of various sizes (Fig. 1F; e.g. plant 2). Surprisingly, one plant (plant 3) was biallelic for two deletions of unexpected sizes, and sequencing revealed that one allele was composed of two small deletions within each sgRNA target, whereas the other allele carried a 140-bp deletion that included most of the target for sgRNA1 and extended beyond the target of sgRNA2 (Fig. 1, F and G).
The remaining T0 plants exhibiting wiry phenotypes were homozygous, biallelic, or chimeric for small insertions and deletions (indels), and sequencing showed that these indels occurred at various positions in proximity to both sgRNA targets. For example, plant 4 was homozygous for two deletions in SlAGO7: a 1-bp deletion located 3 bp upstream of the first PAM site and a 7-bp deletion located 3 bp upstream of the second PAM site (Fig. 1, F and G). Plant 6, on the other hand, was chimeric and contained three alleles, each of which carried a 1-bp deletion in the sgRNA1 target but different small deletions in the sgRNA2 target (Fig. 1, F and G). Finally, cloning and sequencing PCR products from a phenotypically normal T0 plant with an integrated CRISPR/Cas9 T-DNA (plant 8) revealed the expected wild-type allele but also a second allele with a 7-bp deletion in the target of sgRNA1 (Fig. 1, F and G). Together, our results demonstrate that the CRISPR/Cas9 system is highly efficient at generating a range of targeted mutations in stable transgenic tomato plants and that using two sgRNAs allows homozygous deletions of a desired size in the first generation. It should be noted, however, that although the overall rate of mutagenesis was high, a deletion of the expected size in the homozygous state was observed only in one of 29 T0 plants. A possible explanation for this low efficiency is that the DNA cutting at each target must be simultaneous, and the probability of this occurring is significantly lower than asynchronous cuts and repairs at each target, which would result in mutations at each target, as we mostly observed.
A prerequisite for further analysis or application of any mutation is germline transmission and, thus, heritability. We tested whether CRISPR/Cas9-generated mutations from two of our T0 plants could be inherited in the absence of the inductive Cas9/sgRNA1/sgRNA2 transgene. slago7 mutants exhibit poor fertility, but older plants can sometimes produce small amounts of viable pollen (Yifhar et al., 2012). We tested the heritability of two deletions simultaneously by fertilizing wild-type flowers with pollen from plant 3 (Fig. 1H). Although low fertility yielded only four F1 seeds from multiple crosses, PCR genotyping showed that all four progeny plants were heterozygous for either the large or small deletion and the wild-type allele, which we confirmed by sequencing (data not shown). Serendipitously, all four plants lacked the Cas9 transgene, despite an expected 1:1 segregation for a single hemizygous T-DNA insertion (Fig. 1H). We further tested heritability using plant 5, which was biallelic for two small deletions (Fig. 1H). We note that, despite observing a slightly larger PCR product from plant 5, extensive sequencing showed only the two deletion alleles. We hypothesize that this is due to heteroduplex formation during PCR, in which a DNA loop must form in order for different-sized alleles to anneal, resulting in slower migration. PCR genotyping of 10 F1 progeny revealed two plants lacking Cas9: one was heterozygous, carrying the wild-type allele and one of the deletion alleles, whereas the other progeny plant was heterozygous for the second deletion allele (Fig. 1H). Thus, CRISPR/Cas9-induced mutations in tomato can be stably transmitted through the germline.
An important consideration when identifying mutations in regenerated plants carrying the Cas9 transgene is sectoring, as reported in rice (Zhang et al., 2014). In T0 plants and in progeny that still carry Cas9, unless the target sites are no longer present, it cannot be assumed that the genotype detected in a small tissue sample is representative of the entire plant. Additional alleles originating from late-arising chimerism may have been missed in our T0 plants, which could result in different flowers transmitting different alleles. Thus, firm conclusions on the genotypes of whole plants can only be reached by analyzing Cas9-free T1 segregants.
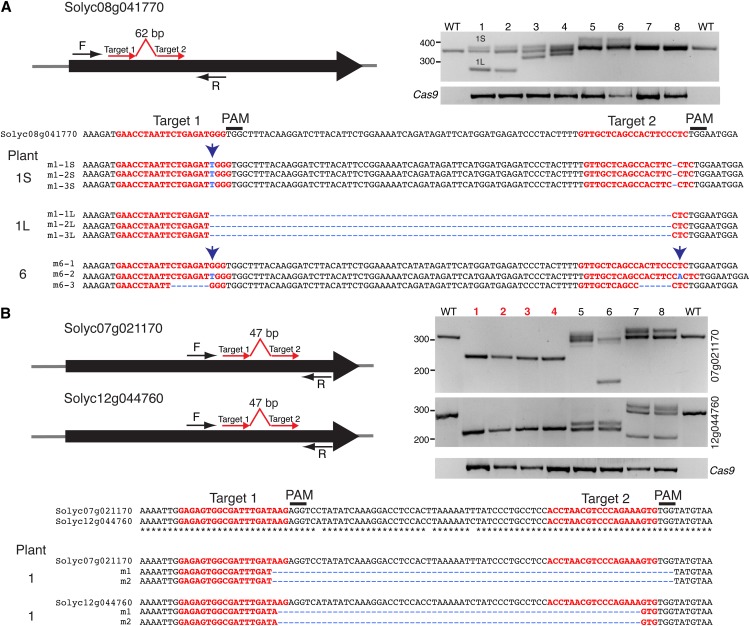
Our results, along with those of others (Feng et al., 2014; Gao and Zhao, 2014; Zhang et al., 2014), suggest that the CRISPR/Cas9 system is poised to rapidly become the technology of choice to generate gene knockouts for reverse genetics studies. As an example, we recently discovered using forward genetics that the gene Solyc11g064850 controls multiple aspects of tomato reproductive development (K. Liberatore, C. MacAlister, and Z.B. Lippman unpublished data). There are three homologs of Solyc11g064850 in the tomato genome (Tomato Genome Consortium, 2012), two of which share high nucleotide identity, suggesting possible redundancy. To test the gene-editing efficiency of CRISPR/Cas9 beyond SlAGO7 and explore its potential for reverse genetics, we generated two additional constructs to target the three homologs of Solyc11g064850. We again incorporated two sgRNAs to generate defined deletions, with one construct designed to target Solyc08g041770 and the other designed to simultaneously target a conserved region in the second to last exon of Solyc07g021170 and Solyc12g044760 (Fig. 2, A and B). For each construct, we regenerated eight T0 plants. PCR genotyping showed that six of eight transgenic lines (75%) recovered from transformation with the CRISPR/Cas9 construct targeting Solyc08g041770 carried mutations, and most plants were likely chimeric (Fig. 2A). For example, PCR genotyping showed that two plants carried the expected deletion, a smaller deletion, and a presumptive wild-type allele. However, sequencing revealed that the latter carried a 1-bp insertion and a 1-bp deletion within the targets of sgRNA1 and sgRNA2, respectively (Fig. 2A, plant 1). Moreover, plant 6, which produced a PCR product dominated by a wild-type size, carried the wild-type allele but also two additional alleles with small indels (Fig. 2A). These results indicate that most plants lacking prominent polymorphisms according to PCR genotyping still harbor mutations. Remarkably, all eight T0 plants carrying the CRISPR/Cas9 construct targeting Solyc07g021170 and Solyc12g044760 showed indels of various sizes (Fig. 2B), and four plants (plants 1–4) carried homozygous deletions in both genes. Sequencing confirmed the expected deletion for Solyc12g044760, whereas the Solyc07g021170 deletion was slightly larger (Fig. 2B). Notably, all four plants originated from the same callus, suggesting that both deletions were induced early in the transformation process, likely before the first division of a single transformed cell. Phenotypic analyses of these plants are ongoing and will be the subject of a future study.
Figure 2.
CRISPR/Cas-induced mutations in a gene family. A, Schematic illustrating two sgRNAs (red arrows) targeting the Solyc08g041770 coding sequence and PCR and sequencing results showing indels in six of eight T0 plants. Plant 1 was chimeric for the desired deletion (1L), another allele (1S) carrying a 1-bp insertion in the target of sgRNA1 (blue arrow), and a 1-bp deletion in the target of sgRNA2. Plant 6 was also chimeric. B, Schematic illustrating two sgRNAs (red arrows) targeting the conserved second to last exon of the Solyc07g021170 and Solyc12g044760 homologs. An alignment of the targeted regions is presented below, and PCR genotyping is shown to the right. Asterisks indicate identical nucleotides. Four of eight T0 plants (red font), which were derived from the same callus, were homozygous for deletions in each gene. Note also the indels of various sizes in four additional chimeric plants.
Recently, the transient use of CRISPR/Cas9 was reported in tomato roots (Ron et al., 2014). Mutations were introduced at desired loci in so-called hairy root structures induced by Agrobacterium rhizogenes carrying a CRISPR/Cas9 transgene. Yet, no transgenic plants were regenerated. To our knowledge, our findings demonstrate for the first time the CRISPR/Cas9-induced mutations in stable transgenic lines of tomato as well as the heritability of these mutations through subsequent generations. Future genome sequencing is needed to assess if, and to what extent, CRISPR/Cas9 causes off targeting in tomato. In the animal field, the CRISPR/Cas9 system was reported to have a high off-target rate (Carroll, 2013). In contrast, several recent studies have shown high specificity of CRISPR/Cas9 in plants (Feng et al., 2014; Gao and Zhao, 2014; Zhang et al., 2014). The issue of off targeting in plants still needs to be addressed systematically, but several approaches can minimize possible impacts, such as using a recently developed algorithm for plant genomes that selects CRISPR/Cas9 sgRNAs with the least predicted off targets (Xie et al., 2014). For reverse genetics studies, choosing a few sgRNAs against a locus that has nonoverlapping off targets can be used to generate multiple independent alleles in different regions of the gene, thereby ensuring that any observed phenotype is caused by mutations in the desired locus rather than by an off target. In any case, backcrossing mutations of interest to wild-type progenitor lines is standard practice in plant genetics, thus mitigating potential off-target effects. When off targeting is of particular concern (e.g. in the case of developing a commercial crop variety), alternative genome editing might be preferred (e.g. TALENs, engineered homing endonucleases). In an accompanying paper, TALENs are shown to be useful for generating heritable mutations in tomato (Lor et al., 2014). However, recent advancements in CRISPR/Cas technology that have already been deployed in nonplant systems, such as the use of double Cas9 nickases (Ran et al., 2013), truncated guide RNAs (Fu et al., 2014), and a fusion of the Flavobacterium okeanokoites (Fok1) nuclease and a catalytically inactive Cas9 (dCas9-Fok1; Guilinger et al., 2014; Tsai et al., 2014), have dramatically reduced off targeting up to 5,000-fold. Ultimately, these improvements will be implemented in plants, providing even more impetus to adopt CRISPR/Cas9 for plant genome editing.
Overall, our findings add to the growing literature demonstrating the power and ease of implementing CRISPR/Cas9 technology across diverse systems and the promise it holds to rapidly advance plant biology and crop improvement.
MATERIALS AND METHODS
CRISPR/Cas9 Construct Design
Constructs were designed to create defined deletions within each target gene coding sequence using two sgRNAs alongside the Cas9 endonuclease gene. The selected sgRNA target sequences were separated by 47 to 68 bp. All constructs were assembled using the Golden Gate cloning method (Weber et al., 2011). Level 1 constructs carrying sgRNAs placed under the control of the Arabidopsis (Arabidopsis thaliana) U6 promoter were assembled as described (Belhaj et al., 2013). Level 1 constructs pICH47732::NOSp::NPTII (Addgene no. 51144 [www.addgene.org]), pICH47742::35S::Cas9 (Addgene no. 49771), pICH47751::AtU6p::sgRNA1, pICH47761::AtU6p::sgRNA2, and the linker pICH41780 (Addgene no. 48019) were assembled into the level 2 vector pAGM4723 (Addgene no. 48015) as described (Weber et al., 2011).
_Agrobacterium tumefaciens_-Mediated Transformation
_A. tumefaciens_-mediated transformations of the standard processing tomato (Solanum lycopersicum) cv M82 were performed according to Van Eck et al. (2006). In brief, cotyledon segments from 6- to 8-d-old seedlings were precultured for 1 d followed by inoculation with A. tumefaciens strain LBA4404 containing the CRISPR/Cas9 constructs of interest. Following a 2-d cocultivation, the cotyledon segments were transferred to a selective regeneration medium that contained 75 mg L−1 kanamycin. When shoots were approximately 1.5 cm tall, they were transferred to a selective rooting medium that also contained 75 mg L−1 kanamycin, and only well-rooted plants were transferred to the greenhouse.
Acclimatization of Transgenic Lines and Greenhouse Conditions
To transfer in vitro plants to greenhouse conditions, the culture medium was thoroughly washed from the roots before transplanting into well-moistened potting soil in plastic pots. They were covered with transparent plastic boxes and maintained in a shaded area of a greenhouse for 5 d. The plastic boxes were removed, and the plants were provided with natural light, supplemented with high-pressure sodium bulbs (50 mmol m−2 s−1) on a 16-h-light/8-h-dark photoperiod. Daytime and nighttime temperatures were 26°C to 28°C and 18°C to 20°C, respectively. The relative humidity was 40% to 60%.
DNA Extraction, PCR Genotyping, and Sequencing
Leaflets were collected from each T0 plant, and genomic DNA was extracted using a standard cetyl-trimethyl-ammonium bromide protocol. Each plant was genotyped for the presence of the Cas9/sgRNA1/sgRNA2 construct with primers designed to amplify a region spanning the 3′ end of the 35S promoter and the 5′ end of Cas9. The 29 transgenic lines positive for integrated CRISPR/Cas9 T-DNA were further genotyped for indel polymorphisms using a forward primer to the left of sgRNA1 and a reverse primer to the right of sgRNA2. All PCR products were resolved on 3% (w/v) agarose gels at 170 V for 80 min. Selected PCR products were excised and purified for cloning into the pSC-A-amp/kan vector (Stratagene). A minimum of five clones per PCR product were sequenced using M13F and M13R primers. Alignments were performed using ClustalW in the MacVector software package.
To test the germline transmission and heritability of CRISPR/Cas9-induced mutations, pollen from plants 3 and 5, which were biallelic for a large and a small deletion and two small deletions, respectively (Fig. 1F), was used to fertilize wild-type flowers. Seeds were germinated and DNA was extracted (cetyl-trimethyl-ammonium bromide) from cotyledons from 1-week-old seedlings. Each seedling was genotyped by PCR for the presence of Cas9 and inheritance of the mutations. PCR products were resolved on a 3% (w/v) agarose gel at 165 V for 85 min. The PCR was purified and cloned into the pSC-A-amp/kan vector, and colonies were sequenced using M13F and M13R primers.
Acknowledgments
We thank members of the Lippman laboratory for discussions, Tim Mulligan and Paul Hanlon for excellent plant care, Jonathan Jones and Sophien Kamoun for reagents, and Weihua Wang and Patricia Keen for help in generating the CRISPR/Cas9 stable transgenic lines. We also thank Yuval Eshed for providing the wiry2-1 image.
Glossary
TALEN
transcription activator-like nuclease
CRISPR
clustered regularly interspaced short palindromic repeats
sgRNA
single guide RNA
PAM
protospacer adjacent motif
T-DNA
transfer DNA
indel
insertions and deletion
Footnotes
1
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. 1237880 to J.V.E. and Z.B.L.).
References
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. (2013) Staying on target with CRISPR-Cas. Nat Biotechnol 31: 807–809 [DOI] [PubMed] [Google Scholar]
- Chen K, Gao C. (2014) Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep 33: 575–583 [DOI] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. (2014) Specific and heritable gene editing in Arabidopsis. Proc Natl Acad Sci USA 111: 4357–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. (2014) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32: 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands AY, Chitwood DH, Plavskin Y, Timmermans MC. (2009) Signals and prepatterns: new insights into organ polarity in plants. Genes Dev 23: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. (2013) TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley MM. (1928) The “wiry” tomato: a recessive mutant form resembling a plant affected with mosaic disease. J Hered 19: 337–344 [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Fan XD. (2014) CRISPR-Cas system: a powerful tool for genome engineering. Plant Mol Biol 85: 209–218 [DOI] [PubMed] [Google Scholar]
- Lor VS, Starker CG, Voytas DF, Weiss D, Olszewski NE. (2014) Targeted mutagenesis of the tomato PROCERA gene using TALENs. Plant Physiol. 166: 1288–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Pennisi E. (2013) The CRISPR craze. Science 341: 833–836 [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, et al. (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646 [DOI] [PubMed] [Google Scholar]
- Van Eck J, Kirk DD, Walmsley AM. (2006) Tomato (Lycopersicum esculentum). Methods Mol Biol 343: 459–473 [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947–951 [DOI] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. (2011) Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE 6: e19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Zhang J, Yang Y. (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant 7: 923–926 [DOI] [PubMed] [Google Scholar]
- Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y. (2012) Failure of the tomato _trans_-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, et al. (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12: 797–807 [DOI] [PubMed] [Google Scholar]