The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulphur proteins (original) (raw)
Abstract
The genome of the yeast Saccharomyces cerevisiae encodes the essential protein Nar1p that is conserved in virtually all eukaryotes and exhibits striking sequence similarity to bacterial iron-only hydrogenases. A human homologue of Nar1p was shown previously to bind prenylated prelamin A in the nucleus. However, yeast neither exhibits hydrogenase activity nor contains nuclear lamins. Here, we demonstrate that Nar1p is predominantly located in the cytosol and contains two adjacent iron–sulphur (Fe/S) clusters. Assembly of its Fe/S clusters crucially depends on components of the mitochondrial Fe/S cluster biosynthesis apparatus such as the cysteine desulphurase Nfs1p, the ferredoxin Yah1p and the ABC transporter Atm1p. Using functional studies in vivo, we show that Nar1p is required for maturation of cytosolic and nuclear, but not of mitochondrial, Fe/S proteins. Nar1p-depleted cells do not accumulate iron in mitochondria, distinguishing these cells from mutants in components of the mitochondrial Fe/S cluster biosynthesis apparatus. In conclusion, Nar1p represents a crucial, novel component of the emerging cytosolic Fe/S protein assembly machinery that catalyses an essential and ancient process in eukaryotes.
Keywords: biosynthesis, cofactor assembly, ferredoxin, H-cluster, mitochondria
Introduction
Hydrogenases are enzymes that either produce or metabolise molecular hydrogen (Peters, 1999; Vignais et al, 2001; Horner et al, 2002). They are usually classified into two major groups based on their metal content. The first group contains iron as the only metal (iron-only hydrogenases), while the second carries nickel plus iron and sometimes additional selenium (NiFe and NiFeSe hydrogenases). Hydrogenases are present not only in Archaea and Bacteria, but also in several eukaryotes such as green algae and protozoa. Surprisingly, a set of close homologues of iron-only hydrogenases is found in virtually all eukaryotic organisms including those that are not known to produce or consume hydrogen such as yeast and man (Horner et al, 2002). These proteins define a family including Saccharomyces cerevisiae Nar1p and human Narf, and they share an overall amino-acid identity of 25–30% (40–50% based on chemical similarity; Figure 1). The sequence similarity of Nar1p-like proteins extends to the ca. 470 C-terminal residues of iron-only hydrogenases such as Desulfovibrio HydA (Nicolet et al, 1999) or clostridial CpI (Peters et al, 1998).
Figure 1.
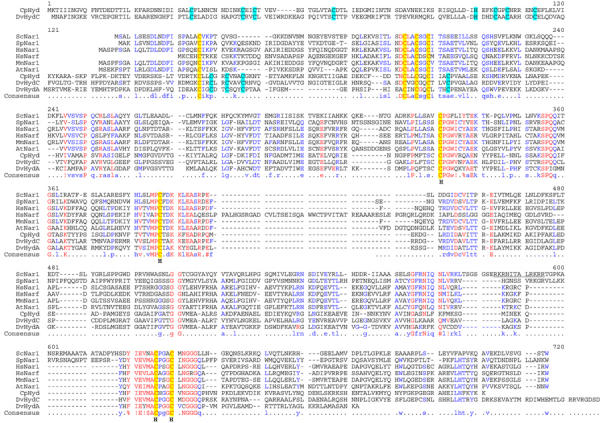
Sequence alignment of Nar1p-like proteins and iron-only hydrogenases. The multisequence alignment of Nar1p-like proteins, Desulfovibrio HydA and HydC, and clostridial CpI hydrogenases was performed using the Multalin program (Corpet, 1988). Conserved cysteine residues are highlighted in yellow and cyan (hydrogenase-specific). The cysteine residues coordinating the H-cluster in iron-only hydrogenases are indicated (H). Abbreviations for organisms: Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe; Hs, Homo sapiens; Mm, Mus musculus; At, Arabidopsis thaliana; Cp; Clostridium pasteurianum; Dv; Desulfovibrio vulgaris.
At its N-terminus, Nar1p carries a ferredoxin-like domain with four conserved cysteine residues that may bind a [4Fe–4S] cluster. The C-terminal part of Nar1p also contains four conserved cysteine residues that, in iron-only hydrogenases, hold a unique H-cluster. This moiety consists of two subclusters, a cubane [4Fe–4S] cluster and a binuclear [2Fe] centre bridged by a cysteine sulphur. The H-cluster forms the catalytic centre of iron-only hydrogenases (Nicolet et al, 2000). Molecular modelling suggests that in Nar1p-like sequences only one side of the cavity containing the H-cluster in iron-only hydrogenases is conserved, which is indicative of a different functional role of Nar1p and its homologues (Nicolet et al, 2002). However, the cellular task of Nar1p-like proteins is unknown to date. So far, only one member of the Nar1p protein family, the human nuclear protein Narf, has been investigated. The protein binds specifically to prenylated prelamin A in the nucleus (Barton and Worman, 1999). However, the functional significance of this interaction has remained unclear, especially since Narf is also present in cells that do not express lamin A. Further, as S. cerevisiae does not contain any lamin homologues, a function of Nar1p in nuclear lamina assembly seems unlikely. Notably, the human genome encodes a second homologue of Nar1p termed Hprn, which exhibits significantly higher similarity to Nar1p and thus is more likely to represent the functional orthologue of Nar1p.
We were interested in Nar1p as a potential nuclear Fe/S protein, since nothing is known to date about the assembly of nuclear Fe/S clusters. In contrast, during the past few years, our knowledge about the biogenesis of mitochondrial and cytosolic Fe/S proteins has increased considerably. The biosynthesis of Fe/S clusters in eukaryotes takes place in mitochondria and is mediated by a complex machinery of at least 10 proteins that are located in the mitochondrial matrix and are derived from the bacterial ancestor of mitochondria (for reviews see Lill and Kispal, 2000; Craig and Marszalek, 2002; Gerber and Lill, 2002; Frazzon and Dean, 2003). A central player of the so-called iron–sulphur cluster (ISC) assembly machinery is the pyridoxalphosphate-dependent cysteine desulphurase Nfs1p, which delivers sulphur for synthesis of Fe/S clusters on the scaffold proteins Isu1p/Isu2p. Electrons are provided for reduction of an unknown substrate by the electron transfer chain consisting of the [2Fe–2S] ferredoxin Yah1p and the ferredoxin reductase Arh1p (Mühlenhoff et al, 2003).
The mitochondrial ISC assembly machinery is also required for maturation of cytosolic Fe/S proteins (Kispal et al, 1999; Lange et al, 2000). To date, three components have been identified that are specifically needed for maturation of cytosolic, but not of mitochondrial, Fe/S proteins, namely the ABC transporter Atm1p of the mitochondrial inner membrane, the intermembrane space protein Erv1p and the tripeptide glutathione (Kispal et al, 1999; Lange et al, 2001; Sipos et al, 2002). A current model suggests that the ABC transporter exports a compound produced by the ISC assembly machinery to the cytosol for Fe/S cluster formation on cytosolic apoproteins. Not much is known about the latter process, but recently a yeast mutant termed cfd1 was described that exhibits a defect in Fe/S cluster assembly of cytosolic, but not of mitochondrial, Fe/S proteins. The CFD1 gene encodes a cytosolic P-loop ATPase, but its precise function is unknown (Roy et al, 2003). A homologous protein called ApbC has been shown to be required for Fe/S cluster-dependent metabolism in Salmonella enterica (Skovran and Downs, 2003). Further, the nitrogenase Fe protein NifH, a distant Cfd1p homologue, is also involved in biosynthesis and insertion of the FeMo cofactor into nitrogenase MoFe protein (Robinson et al, 1987). Together, these observations suggest an important role of P-loop ATPases in Fe/S cluster assembly.
In this report, we show that yeast Nar1p is an essential Fe/S protein located in the cytosol where part of it is associated with membranes. Fe/S cluster assembly on Nar1p is dependent on the mitochondrial ISC machinery. Surprisingly, depletion of Nar1p in a regulatable yeast mutant leads to severe defects in Fe/S cluster assembly on cytosolic, but not on mitochondrial, Fe/S proteins. Moreover, Nar1p was required for maturation of a nuclear Fe/S protein. Hence, our studies for the first time define a function of the hydrogenase-like Nar1p and possibly its eukaryotic homologues in the biosynthesis of extramitochondrial Fe/S proteins.
Results
Nar1p is an Fe/S protein
The close similarity of eukaryotic Nar1p-like proteins to bacterial hydrogenases suggested that Nar1p is an Fe/S protein containing two Fe/S prosthetic groups (Horner et al, 2002; Nicolet et al, 2002). To investigate this, a _Strep_-tagged protein was overproduced in Escherichia coli and purified by _Strep_-Tactin affinity chromatography. Recombinant Nar1p as isolated was yellowish-brown, and the visible absorption spectrum showed a broad, unstructured ‘shoulder' around 420 nm (Figure 2A) typical for [4Fe–4S]2+ clusters and no structured features usually found for [2Fe–2S]2+ clusters (Orme-Johnson and Orme-Johnson, 1982). Upon reduction with 1 mM sodium dithionite, partial bleaching of the absorbance in the visible range occurred.
Figure 2.
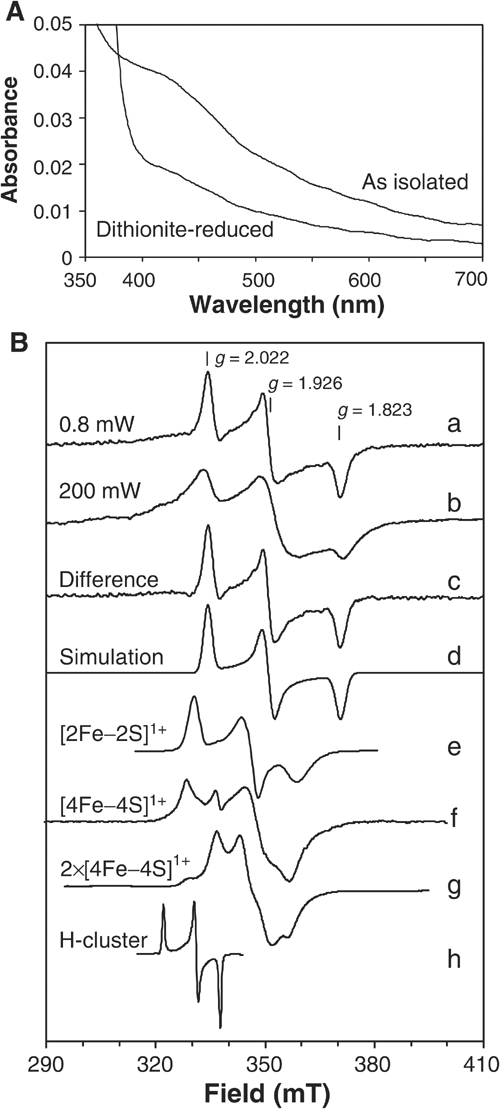
Spectroscopic analysis of purified Nar1p. (A) Absorption spectra of _Strep_-tagged Nar1p (1.6 μM) as isolated and after reduction with 1 mM sodium dithionite in buffer TNE (100 mM Tris–HCl, pH 8, 150 mM NaCl, 1 mM EDTA) containing 40 μM desthiobiotin. (B) EPR spectra of recombinant Nar1p were recorded under the following conditions: microwave frequency, 9.460±0.001 GHz; modulation frequency, 100 kHz; modulation amplitude, 1.25 mT. (a) Nar1p (50 μM) reduced with 5 mM sodium dithionite in buffer TNE with 1 mM desthiobiotin (0.80 mW, 10 K). (b) As in (a), but recorded at 200 mW, 10 K. (c) The sharp rhombic EPR signal obtained by subtracting an appropriate intensity of spectrum b from a. (d) A theoretical simulation of spectrum c (Beinert and Albracht, 1982). Simulation parameters, g _z_=2.022, g _y_=1.926 and g _x_=1.823; linewidths 2.5, 3.0 and 3.1 mT, respectively. For comparison, traces (e–h) were added with appropriate corrections of the magnetic fields to the microwave frequency of Nar1p experiments. (e) [2Fe–2S]1+ cluster in reduced spinach leaf ferredoxin (Hagen and Albracht, 1982). (f) [4Fe–4S]1+ cluster in partially dithionite-reduced heterodisulphide reductase from Methanothermobacter marburgensis (9.459 GHz, 20 mW, 30 K; courtesy of Dr R Hedderich (MPI for Terrestrial Microbiology, Marburg). (g) Two interacting [4Fe–4S]1+ clusters in fully reduced ferredoxin from Acidaminococcus fermentans (9.65 GHz, 2 mW, 12 K; Thamer et al, 2003). (h) Oxidised H-cluster in thionine-treated C. pasteurianum Fe-hydrogenase CpI (9.23 GHz, 10 mW, 20 K; Adams, 1987).
To further substantiate the presence of Fe/S clusters in recombinant Nar1p, low-temperature electron paramagnetic resonance (EPR) spectroscopy was performed. The dithionite-reduced sample gave a rhombic EPR signal with g values of g _z_=2.022, g _y_=1.926 and g _x_=1.823 (Figure 2B, trace a). This EPR signal was untypical compared to clusters of well-characterised Fe/S proteins. For instance, the g values and line shapes did not reflect typical [4Fe–4S]1+ clusters and did not conform to [2Fe-2S]1+ clusters (cf. Figure 2B; traces e–g). Moreover, the signal was distinct from that reported for the H-cluster of iron-only hydrogenases (trace h). Notably, the H-cluster is EPR-active in the oxidised form only (Pierik et al, 1992), whereas Nar1p was EPR-silent in the oxidised (as isolated) form and required reduction for generating EPR signals. The EPR signal disappeared above 30 K (not shown). This finding together with the absorption spectrum (Figure 2A) suggested that Nar1p contained no [2Fe–2S] clusters (Orme-Johnson and Orme-Johnson, 1982; also see below). At higher power, the rhombic EPR signal saturated (_P_1/2=5 mW) and an underlying, broad signal became evident (trace b). The spectral contribution of the rhombic signal obtained by subtraction of the broad signal from trace a (trace c) could be theoretically simulated (trace d) and accurately matched with the experimental spectrum (Beinert and Albracht, 1982).
Broad EPR signals are typical for magnetically coupled Fe/S clusters. In bacterial ferredoxins and iron-only hydrogenases, two [4Fe–4S]1+ clusters at 1–1.5 nm distance are known to give rise to broad, magnetically coupled EPR spectra, which are not just the sum of two individual [4Fe–4S]1+ clusters (Figure 2B, trace g; Mathews et al, 1974). Thus, we suggest that the broad, less-featured EPR signal of Nar1p was derived from magnetic interaction between the rhombic EPR signal and another cluster in a fully reduced state. This interpretation was supported by the observation of the broad signal upon photoreduction using 5′-deazaflavin, which led to complete reduction of Nar1p (not shown). The observed average g value of the coupled spectrum (1.91; Figure 2B, trace b) is built up from the average g value of the coupling partner (calculated to be 1.90) and that of the rhombic signal (1.92; trace a). The coupling partner therefore most likely also represents an Fe/S cluster, an idea that is consistent with the visible absorption spectrum (Figure 2A) and the eight conserved cysteine residues in Nar1p-like proteins (Figure 1).
Chemical analysis of the _Strep_-tagged protein purified under anaerobic conditions revealed the presence of 3.5±0.2 atoms of non-haeme iron and 2.9±0.1 atoms of acid-labile sulphur per protein. The amount of Fe and S in Nar1p was less than expected from the existence of two clusters detected by EPR. We assume that during overproduction in E. coli only a fraction of Nar1p was converted to an Fe/S holoprotein. From earlier studies on nitrogenase, it is known that multiple Fe/S clusters may be integrated into apoproteins either completely or not at all (Smith et al, 1980). Taken together, our initial spectroscopic characterisation demonstrates that Nar1p is a complex Fe/S protein. Most likely, it contains two coupled Fe/S clusters, one of them being an unusual Fe/S cluster.
Depletion of Nar1p leads to growth arrest, which cannot be restored by expression of human Nar1p homologues
To initiate functional studies on Nar1p, we constructed a regulatable mutant of NAR1. To this end, 197 nucleotides upstream of the NAR1 start codon were replaced with the GAL1-10 promoter by homologous recombination to yield strain Gal-NAR1. When Gal-NAR1 cells were grown in medium containing galactose, Nar1p was produced at higher levels than in wild-type cells (Figure 3A), but cell growth was unaffected (Figure 3B). When glucose was used as the sole carbon source in order to deplete Nar1p, Nar1p-specific bands could not be detected in cell lysates by immunostaining, confirming the specificity of our antiserum (Figure 3A). Gal-NAR1 cells did not give rise to colonies on glucose-containing agar plates (Figure 3B), showing that NAR1 is indispensable for viability of yeast cells.
Figure 3.
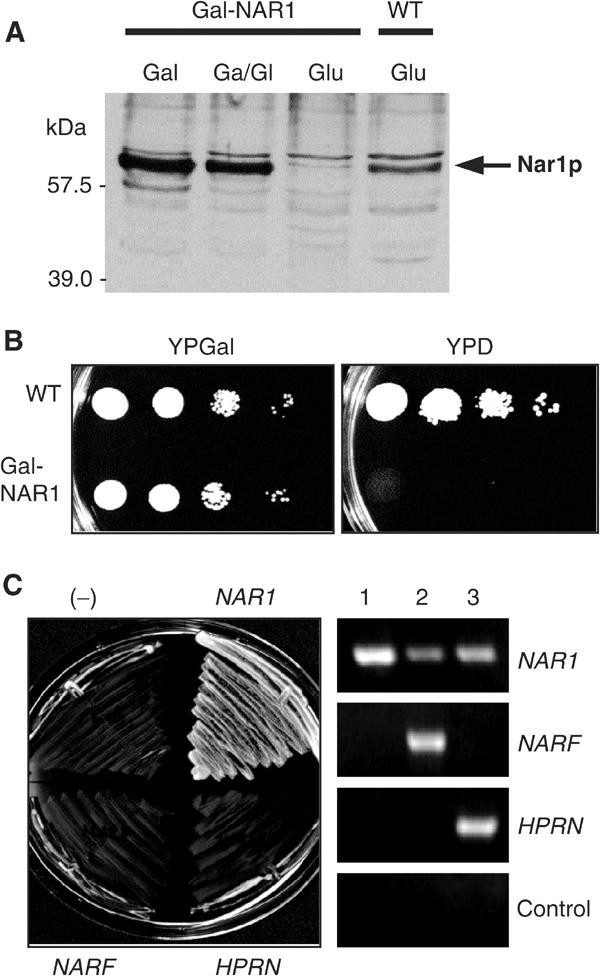
Yeast NAR1 is an essential gene and cannot be replaced by its human homologues. (A) Wild-type (WT) and Gal-NAR1 cells were grown in liquid rich medium supplemented with galactose (Gal), a 1:1 mixture of galactose and glucose (Ga/Gl) or glucose (Glu). A total cell extract prepared by alkaline lysis was subjected to immunostaining for Nar1p using an anti-Nar1p antiserum. The minor band at 57 kDa may represent a breakdown product of mature Nar1p. (B) Wild-type and Gal-NAR1 cells were grown for 2 × 2 days at 30°C on agar plates containing rich media supplemented with galactose (YPGal) or glucose (YPD). 10-fold serial dilutions are shown. (C) Gal-NAR1 cells were transformed with plasmid p416MET25 containing either no insert (−), the yeast NAR1 gene, or the genes of human NARF and HPRN. Cells were grown on YPD medium for 2 × 2 days at 30°C (left). Expression of the human genes was tested by RT–PCR using galactose-grown cells harbouring plasmids with (1) yeast NAR1, (2) NARF and (3) HPRN genes. A control reaction was performed without reverse transcriptase.
To address whether the human homologues Narf and Hprn can functionally replace Nar1p, the corresponding genes were expressed in Nar1p-depleted Gal-NAR1 cells (Figure 3C, right panel). Neither of the human genes was capable of rescuing the lethal phenotype of Gal-NAR1 cells upon growth on glucose-containing medium (left panel). On the contrary, yeast NAR1 fully restored the growth defect of Gal-NAR1 mutant cells showing that these cells specifically lack Nar1p under these conditions. We conclude that the human Nar1p-like proteins cannot replace the cellular task of Nar1p in yeast. Nevertheless, the lack of complementation of Nar1p-deficient yeast cells by the human genes does not exclude that these proteins may fulfil orthologous functions in yeast and man.
Nar1p is predominantly localised in the cytosol
The subcellular localisation of Nar1p was studied by in situ immunofluorescence. Yeast cells overexpressing NAR1 were labelled using an affinity-purified antibody followed by incubation with a fluorophore-conjugated secondary antibody. The immunofluorescence was distributed in a similar fashion as that found after labelling with monoclonal antibodies against Pgk1p (phosphoglycerate kinase), a cytosolic marker protein (Figure 4A). The signal was specific for overproduced Nar1p, because no fluorescence was detectable in wild-type cells. Apparently, the endogenous levels of Nar1p were too low for detection by our immunostaining procedure. This result was in good agreement with the low expression levels of NAR1 in wild-type cells as determined by our immunoblot analysis and by systematic protein expression analysis (Figure 3A; Ghaemmaghami et al, 2003).
Figure 4.
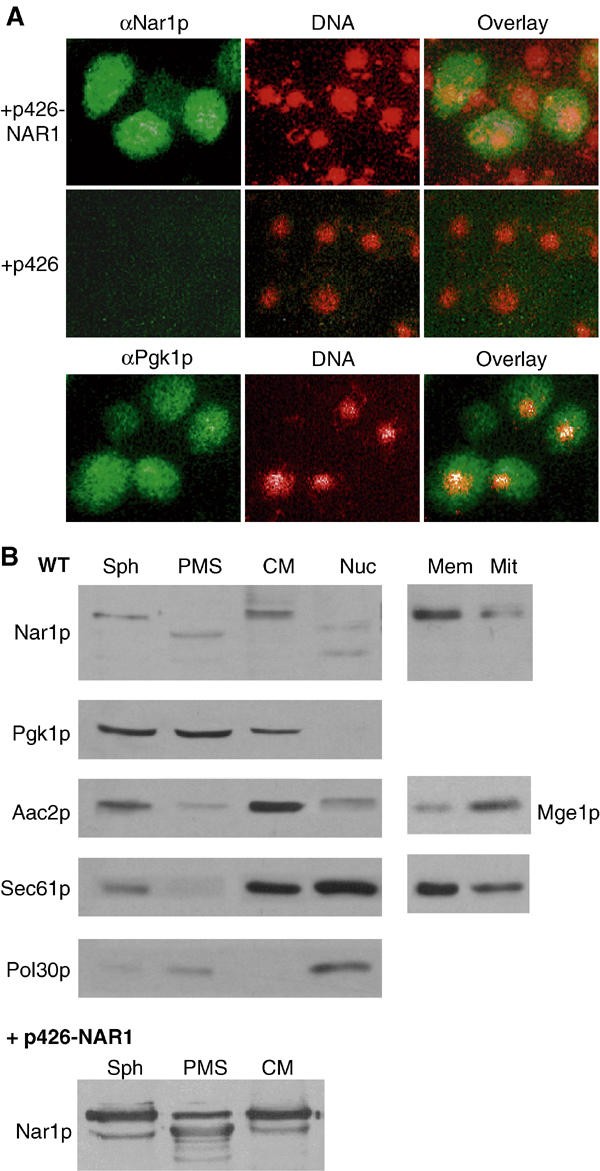
Subcellular localisation of Nar1p. (A) In situ localisation of Nar1p. Wild-type cells (strain PSY581) were transformed with the high-copy expression vector p426 containing NAR1 or no gene. Log-phase cells were fixed with 2.4% (w/v) formaldehyde, permeabilised and labelled with purified anti-Nar1p (αNar1p) or monoclonal anti-Pgk1p (αPgk1p) antibodies, followed by fluorophore-conjugated secondary antibodies (green). DNA was counterstained with DAPI and is indicated in red. Similar results with low fluorescence intensity were obtained with NAR1 inserted into low-copy expression vector p416MET25. (B) Immunoblot analysis of Nar1p in cell fractions. Log-phase wild-type cells were converted into spheroplasts (Sph), subjected to Dounce homogenisation and, after removal of intact cells and cell debris, the extract was fractionated into post-mitochondrial supernatant (PMS) and crude mitochondria (CM) by centrifugation for 10 min at 12 000 g. A similar fractionation was performed with cells overproducing Nar1p from vector p426-NAR1. A separate wild-type yeast culture was used to isolate nuclei (Nuc) by Ficoll gradient centrifugation (Aris and Blobel, 1991). Equal amounts of protein (20 μg/per lane) were separated by SDS–PAGE, blotted and immunostained for Nar1p or marker proteins of known cellular localisation (left panel; cytosolic phosphoglycerate kinase Pgk1p; mitochondrial ATP/ADP carrier Aac2p; endoplasmic reticulum translocon subunit Sec61p; and nuclear DNA polymerase-associated Pol30p). The crude mitochondrial fraction was further separated on a Nycodenz step-gradient into fractions containing enriched mitochondria (Mit) and microsomal membranes (Mem). Samples were analysed by immunostaining (right panel; Mge1p; mitochondrial matrix).
Because overexpression can occasionally lead to artefacts, we used subcellular fractionation of wild-type and Nar1p-overproducing cells as an independent method for Nar1p localisation. Cells were converted into spheroplasts, lysed, separated by centrifugation into post-mitochondrial supernatant and pellet fractions, and the fractions were analysed by protein blot analysis. Endogenous and overproduced Nar1p behaved similarly and were found in the post-mitochondrial supernatant fraction (ca. 80% of total) as well as in the pellet fraction containing crude mitochondria and other organelles (Figure 4B, left). In the supernatant fraction, a band of smaller mass was found, which, based on its immunoreactivity and expression pattern, is a degradation product of Nar1p. Degradation was seen even when the procedure was performed in the presence of protease inhibitors and under anaerobic conditions (not shown). Apparently, Nar1p is rather sensitive to proteolysis. When the crude mitochondrial fraction was further separated by density-gradient centrifugation, Nar1p was enriched in the microsomal membrane fraction (Figure 4B, right). In contrast, effective depletion of Nar1p in purified mitochondria was achieved excluding that Nar1p is associated with these organelles. In Ficoll-purified nuclei, Nar1p-specific bands of lower mass, but no full-length Nar1p, were detected. These bands also may be degradation products generated during the lengthy isolation procedure. Taken together, these data indicate that Nar1p is localised predominantly in the cytosol and part of it is membrane-associated. The localisation of Nar1p is similar to Cfd1p (Roy et al, 2003), but clearly differs from that of its human homologue Narf, which was found to be exclusively nuclear (Barton and Worman, 1999).
The mitochondrial ISC machinery is required for Fe/S cluster maturation of Nar1p
The mitochondrial ISC assembly and export machineries were previously demonstrated to be required for Fe/S protein maturation in the cytosol (Lill and Kispal, 2000). We therefore tested whether the de novo maturation of the Nar1p Fe/S protein depends on components of the mitochondrial ISC assembly and export pathways. We took advantage of cells in which various mitochondrial ISC components such as the cysteine desulphurase Nfs1p, the ferredoxin Yah1p and the ABC transporter Atm1p can be depleted by the use of the GAL1-10 promoter upstream of the respective ISC gene (Kispal et al, 1999; Lange et al, 2000). These mutant cells and wild-type cells were transformed with a plasmid for overexpression of NAR1, and were cultured in low-iron medium containing galactose or glucose followed by radiolabelling with 55Fe. After lysis of the cells, Nar1p was immunopurified from the cell extract using a specific anti-Nar1p antiserum. Nar1p was the major protein isolated in this way as verified by silver staining (not shown). The assembly of Fe/S clusters was estimated from the 55Fe incorporation into Nar1p by scintillation counting (Kispal et al, 1999). In wild-type cells or in galactose-grown Gal-NFS1, Gal-YAH1 and Gal-ATM1 cells, that is, cells that synthesised the respective Isc proteins, a significant amount of 55Fe was incorporated into Nar1p (Figure 5A, left bars). In cells that did not overproduce Nar1p, three- to five-fold lower amounts of radioactivity were found (Figure 5A, third bars). Since endogenous Nar1p was not detectably associated with the immunobeads (not shown), these amounts represented the background of our assay.
Figure 5.
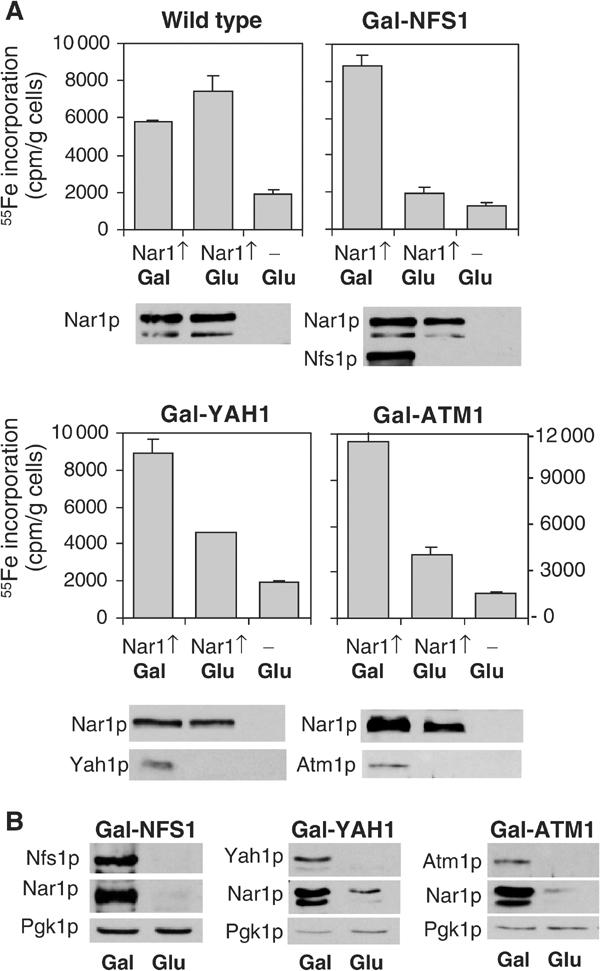
Fe/S cluster assembly on Nar1p depends on the mitochondrial ISC machinery. (A) Wild-type and the galactose-regulatable Gal-NFS1, Gal-YAH1 and Gal-ATM1 cells were transformed with plasmid p426-NAR1 (Nar1↑) or empty vector p426 (−). Cells were grown in iron-poor medium supplemented with galactose (Gal) or glucose (Glu) for 24 h. Cells were radiolabelled with 55Fe and disrupted with glass beads in Triton X-100-containing buffer. Nar1p was isolated from the cell extract by immunoprecipitation and the amount of 55Fe associated with Nar1p was quantified by scintillation counting. The indicated proteins were visualised by immunostaining of cell extracts. (B) The indicated cells overproducing Nar1p were grown for 40 h and cell extracts were analysed as in (A).
Upon depletion of Nfs1p, Yah1p or Atm1p by growth of the respective cells in glucose-supplemented medium for 24 h, the amount of 55Fe associated with the anti-Nar1p immunobeads was largely diminished as compared to the wild-type situation, suggesting a requirement of the mitochondrial ISC components for maturation of Nar1p (Figure 5A, second bars). At this time point, growth rates were like that of wild-type cells and iron uptake into the cells was not limiting, but rather somewhat higher than that of wild-type controls (Kispal et al, 1999; Lange et al, 2000). The decrease in 55Fe incorporation into Nar1p was not due to the use of glucose per se because in wild-type cells the carbon source made little difference for 55Fe incorporation into Nar1p (Figure 5A). Further, the Nar1p protein levels were similar in Gal-NFS1 and Gal-YAH1 cells under both growth conditions, indicating that a large fraction of Nar1p was in the apoform upon depletion of Nfs1p and Yah1p. In Atm1p-depleted cells, however, the level of Nar1p was diminished, approximately correlating with the decrease in 55Fe associated with the immunobeads. We presume that the Nar1p apoprotein became readily susceptible to proteolysis in Atm1p-depleted cells, a behaviour that is often observed for Fe/S apoproteins (Kispal et al, 1999; Chen et al, 2002). In support of this notion, substantial degradation of Nar1p was also observed in Gal-NFS1 and Gal-YAH1 cells upon growth for extended times (40 h) in glucose-supplemented medium (Figure 5B). The effect was not due to general proteolysis in glucose-cultured cells, since the levels of cytosolic phosphoglycerate kinase (Pgk1p) were similar for both carbon sources. Rather, degradation of Nar1p seemed to be a consequence of the highly unstable nature of the apoform of Nar1p. Taken together, the insertion of Fe/S clusters into Nar1p requires components of the mitochondrial ISC assembly and export machineries. In turn, this dependence supports our finding that Nar1p is an Fe/S protein.
Nar1p is essential for Fe/S cluster assembly on cytosolic, but not on mitochondrial, Fe/S proteins
In the course of our search for the cellular function of Nar1p, we considered its participation in the biogenesis of cytosolic and nuclear Fe/S proteins. To address this possibility, we first analysed the enzyme activity of isopropylmalate isomerase (Leu1p), a cytosolic Fe/S protein (Kispal et al, 1999). Wild-type and Gal-NAR1 cells were grown for 40 h in minimal medium supplemented with either galactose or glucose to induce or repress, respectively, synthesis of Nar1p in Gal-NAR1 cells. When Nar1p was depleted, the Leu1p enzyme activity was diminished more than eight-fold, while no effect was seen in wild-type cells (Figure 6A). Since the protein level of Leu1p was unchanged upon depletion of Nar1p (inset), this result is a first indication that Nar1p may be required for maturation of Leu1p.
Figure 6.
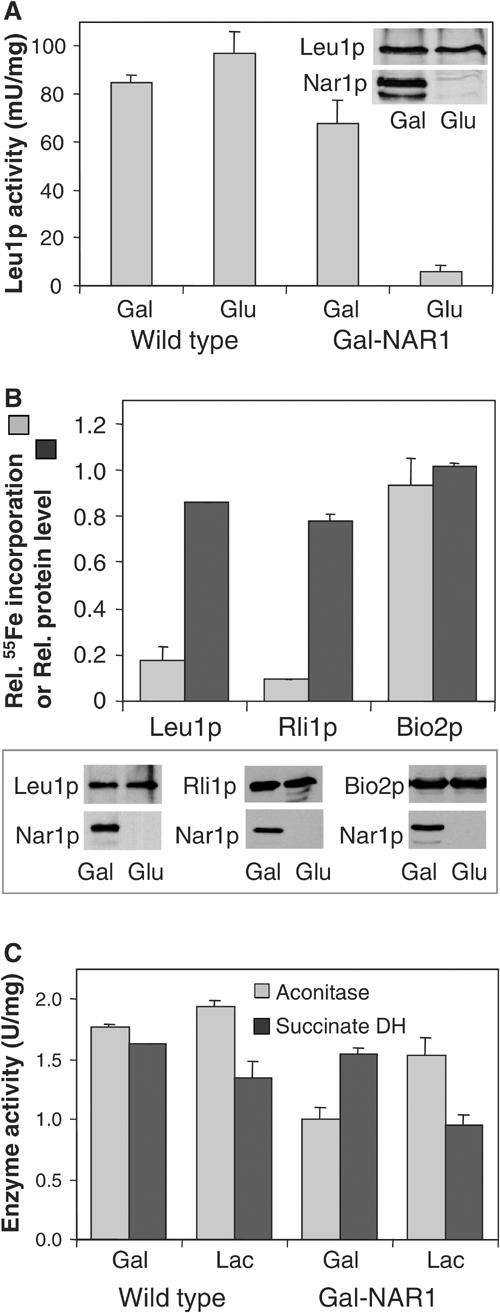
Nar1p is required for maturation of cytosolic, but not of mitochondrial, Fe/S proteins. (A) Wild-type and Gal-NAR1 cells were grown in minimal medium supplemented with galactose (Gal) or glucose (Glu). The enzyme activity of the cytosolic isopropylmalate isomerase (Leu1p) was estimated in total cell extracts. The levels of Leu1p and Nar1p in Gal-NAR1 cells were measured by immunoblot analysis (inset). (B) Gal-NAR1 cells were grown in iron-poor medium supplemented with galactose or glucose. Cells were transformed with high-copy plasmids carrying the genes for an HA-tagged version of cytosolic Rli1p or for mitochondrial Bio2p. Radiolabelling with 55Fe, preparation of cell extracts and immunoprecipitation of the Fe/S protein of interest were performed as in Figure 5. For Leu1p, the endogenous protein was analysed. Results are given as the ratio of 55Fe incorporation in Nar1p-depleted (glucose-grown) cells and that in Nar1p-expressing (galactose-grown) cells, corrected for a small background (<2% of total signal of galactose-grown cells). Protein levels of the indicated proteins were determined by immunoblot analysis (lower panel) and quantified by densitometry (upper panel). (C) The enzyme activities (in U per mg of mitochondrial protein) of aconitase and succinate dehydrogenase (DH) were assayed in mitochondria isolated from wild-type or Gal-NAR1 cells after growth in minimal medium containing galactose or lactate (Lac).
The loss of Leu1p enzyme activity may be due to damage or impaired synthesis of the Fe/S cofactor. To discriminate between these alternatives, the de novo Fe/S cluster maturation of this Fe/S protein was analysed in Gal-NAR1 cells using the 55Fe radiolabelling assay described above. Radiolabelled iron associated with Leu1p was decreased by 80% upon depletion of Nar1p, while the Leu1p protein levels were down by only 15% (Figure 6B). Similar results were obtained for a second cytosolic Fe/S protein, Rli1p (Figure 6B; Lange et al, 2001). These results demonstrate that Nar1p is required for the de novo maturation of cytosolic Fe/S proteins. In turn, the findings suggest that the loss of Leu1p enzyme activity in Nar1p-depleted cells was due to a defect in Fe/S cluster assembly rather than caused by Fe/S cluster damage.
We next tested whether depletion of Nar1p affected the assembly of mitochondrial Fe/S proteins. Gal-NAR1 cells were grown for 40 h in minimal medium containing 0.1% (w/v) glucose and the nonfermentable carbon source lactate in order to deplete Nar1p while maintaining mitochondrial biogenesis. Under these growth conditions, Leu1p activity was as strongly diminished as after growth of Gal-NAR1 cells in the presence of glucose (not shown; cf. Figures 6A and B). The enzyme activities of the mitochondrial Fe/S proteins aconitase and succinate dehydrogenase (complex II) in isolated mitochondria were hardly affected by depletion of Nar1p (Figure 6C). Further, we investigated the de novo 55Fe/S cluster assembly into the mitochondrial Fe/S protein Bio2p. The 55Fe incorporation into Bio2p was not decreased upon depletion of Nar1p, in contrast to the findings made for cytosolic Fe/S proteins (Figure 6B). Together, these results clearly demonstrate that depletion of Nar1p results in a severe defect of Fe/S cluster assembly on cytosolic, but not on mitochondrial, Fe/S proteins. The finding is consistent with the fact that Nar1p was not associated with purified mitochondria.
Nar1p is involved in the maturation of a nuclear Fe/S protein
Since nothing is known about the biogenesis of nuclear Fe/S proteins, we chose the N-glycosylase Ntg2p as a nuclear Fe/S marker protein to study its maturation (Alseth et al, 1999). An HA-tagged version of Ntg2p was overproduced in Gal-NAR1 cells, and the exclusive nuclear localisation of the overproduced HA-tagged Ntg2p was verified by immunofluorescence (Figure 7A). Maturation of HA-tagged Ntg2p was assayed by 55Fe radiolabelling and immunoprecipitation as described above. Upon depletion of Nar1p, the amount of 55Fe associated with Ntg2p was diminished more than 10-fold as compared to Nar1p-containing cells, even though almost wild-type levels of Ntg2p polypeptide were detectable by immunostaining (Figure 7B). We conclude from these findings that Nar1p is also required for the maturation of nuclear Fe/S proteins.
Figure 7.
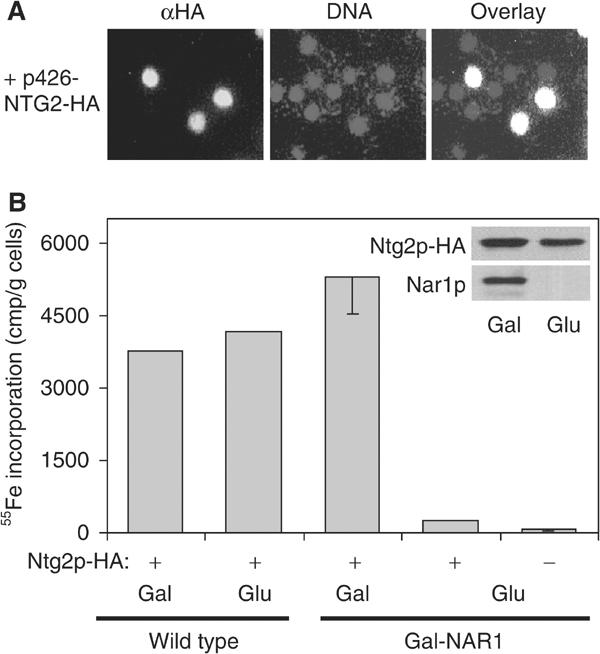
Maturation of the nuclear Fe/S protein Ntg2p depends on Nar1p function. Wild-type or Gal-NAR1 cells were transformed with a high-copy plasmid (p426-NTG2-HA) encoding an HA-tagged version of Ntg2p. (A) Localisation of overproduced HA-tagged Ntg2p by immunofluorescence was performed as in Figure 4A. (B) After growth of cells in galactose- (Gal) or glucose-containing (Glu) iron-poor minimal medium, maturation of Ntg2p was measured by following the 55Fe incorporation by immunoprecipitation with anti-HA antibodies as described in Figure 5A. A control immunoprecipitation was performed with cells lacking HA-tagged Ntg2p (−). The inset shows immunostaining of Ntg2p-HA and Nar1p in extracts of Gal-NAR1 cells.
Nar1p-depleted cells do not accumulate iron in mitochondria
The mitochondrial Fe/S cluster assembly mutants investigated to date display a characteristic alteration in cellular iron homeostasis (see Gerber and Lill, 2002). Iron uptake into these cells is increased substantially, correlating with the constitutive expression of the Aft1p/Aft2p transcription factor-dependent Fe regulon (Foury and Talibi, 2001). The excess iron is accumulated in mitochondria (see, e.g., Kispal et al, 1997). Are similar effects observed upon inactivation of Nar1p? To address this question, Nar1p and, as a control, Atm1p were depleted in Gal-NAR1 and Gal-ATM1 cells, respectively, by growth in lactate media (not shown), and mitochondria were isolated. The total amount of iron associated with mitochondria isolated from Nar1p-depleted cells was not significantly higher than that of wild-type cells or Nar1p-containing Gal-NAR1 cells (Figure 8). In contrast, depletion of Atm1p led to a dramatic accumulation of Fe in mitochondria (Kispal et al, 1997). Thus, even though depletion of Atm1p and Nar1p exhibits similar defects in the assembly of cytosolic Fe/S proteins, Nar1p-depleted cells did not show any iron accumulation in mitochondria. Likewise, we did not detect any major changes in the cellular iron uptake efficiency upon Nar1p depletion (not shown). We conclude that the absence of Nar1p does not cause the dramatic alterations in iron homeostasis that are observed upon depletion of proteins of the mitochondrial ISC biogenesis machinery including Atm1p.
Figure 8.
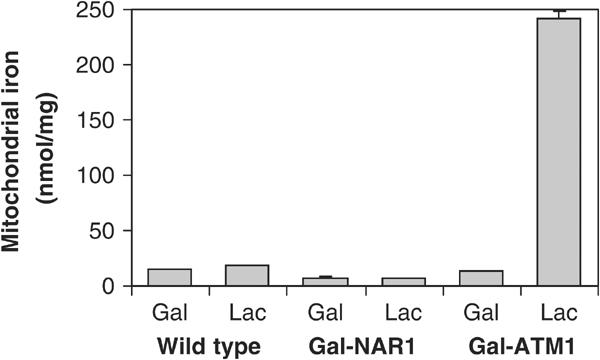
Depletion of Nar1p does not lead to iron accumulation in mitochondria. Wild-type, Gal-NAR1 and Gal-ATM1 cells were grown for 40 h in minimal medium containing galactose (Gal) or in lactate medium (Lac). Mitochondria were isolated and the amount of total iron was determined.
Discussion
In this communication, we provide the first functional characterisation of a member of the Nar1 protein family. These proteins are found in virtually all eukaryotes and show striking sequence similarity to bacterial iron-only hydrogenases, but are not known to function as hydrogenases. We show that yeast Nar1p is an essential, cytosolic Fe/S protein containing an unusual Fe/S cluster interacting with a second Fe/S cluster. Biogenesis of these Fe/S clusters strictly depends on the function of the mitochondrial ISC assembly and export machineries. In vivo studies employing a regulatable NAR1 mutant cell show that Nar1p performs a crucial function in the biogenesis of both cytosolic and nuclear, but not of mitochondrial, Fe/S proteins. Given its essence for viability and the conservation of the Fe/S cluster assembly pathway, participation in Fe/S protein biogenesis may be the basic function of all members of the Nar1 protein family. Nar1p appears to be a novel component of the poorly characterised biogenesis apparatus for Fe/S proteins in the eukaryotic cytosol. The only other known constituent of this pathway is the cytosolic P-loop ATPase Cfd1p (Roy et al, 2003). It will be interesting to investigate whether these two proteins interact, genetically or directly, by protein–protein association.
Even though the presence of iron-only hydrogenase homologues in eukaryotes that do not metabolise hydrogen has been noted before (e.g., Vignais et al, 2001; Horner et al, 2002; Nicolet et al, 2002), the functional role of this protein family has remained enigmatic. Our demonstration of an involvement of yeast Nar1p in the biogenesis of cytosolic and nuclear Fe/S proteins relies on a defect in the enzyme activity of the cytosolic Fe/S protein Leu1p and the striking impairment of 55Fe incorporation into cytosolic and nuclear Fe/S model proteins. Mitochondrial Fe/S proteins on the other hand exhibited normal activity and showed wild-type efficiencies of Fe/S cluster assembly. These findings exclude that depletion of Nar1p evoked conditions in the cell that lead to a general damage of Fe/S clusters, for example, by oxidative stress. Rather, Nar1p appears to be required for de novo maturation of cytosolic and nuclear Fe/S proteins. Since Nar1p was shown here to represent an Fe/S protein itself, these data imply that Nar1p is involved in its own maturation. A similar ‘chicken-and-egg' situation has been described previously for the biogenesis of the mitochondrial ferredoxin Yah1p, in that assembly of its [2Fe–2S] cluster depends on the presence of the mitochondrial ISC machinery (Lange et al, 2000). Yah1p is a central component of this machinery, which, in conjunction with ferredoxin reductase Arh1p, supplies electrons for Fe/S cluster assembly on the scaffold proteins Isu1p/Isu2p (Mühlenhoff et al, 2003).
At present, we can only speculate about the detailed molecular function of Nar1p in Fe/S protein maturation. Like Yah1p, Nar1p may also perform an electron transfer function, since the homologous iron-only hydrogenases catalyse the electron shuttling to protons in Bacteria (e.g., Clostridium, Desulfovibrio), in hydrogenosomes of eukaryotic parasites (Trichomonads, cytrid fungi) and in chloroplasts of green algae (Chlamydomonas, Chlorella; Horner et al, 2002). It is thus tempting to speculate that in the eukaryotic cytosol the electron transfer function of hydrogenase-related Nar1 proteins was utilised for a novel task, namely Fe/S protein biogenesis. In vitro reconstitution of Fe/S protein maturation in the eukaryotic cytosol and the identification of further components of this pathway will now be necessary to gain insights into the molecular mechanism of this process and into the particular function of Nar1p.
A characteristic and general phenotype of mitochondrial mutants in Fe/S protein assembly is the accumulation of iron in mitochondria (Gerber and Lill, 2002). The resulting induction of the Aft1p/Aft2p transcription factor-dependent iron regulon leads to increased cellular iron uptake (Foury and Talibi, 2001; Rutherford et al, 2001). Mutants in the ISC export components Atm1p, Erv1p and in glutathione synthesis also show this behaviour (Kispal et al, 1999; Lange et al, 2001; Sipos et al, 2002). Since these mutants, as Nar1p-depleted cells, display defects only in cytosolic, but not in mitochondrial, Fe/S proteins, it was a surprise to us that Nar1p-deficient cells did not show any immediate alterations in their cellular iron metabolism. Neither the uptake of iron into the cell nor the intracellular distribution was significantly changed. Apparently, yeast mitochondria serve as a master control device for cellular iron homeostasis. For iron regulation, the organelles seem to use a product of the Fe/S protein metabolism, but not haeme, the other major iron-containing compound synthesised by mitochondria. This behaviour is notably different from human cells, which accumulate iron not only after Fe/S protein biogenesis defects, but also as a result of impaired porphyrin metabolism (Bekri et al, 2000; Cazzola et al, 2003). Despite the less complex regulation of iron uptake and distribution in yeast cells, our current study suggests that, under the assumption of a direct function of Nar1p in maturation of all extramitochondrial Fe/S proteins, a cytosolic or nuclear Fe/S protein is unlikely to be involved in this regulatory circuit. Our findings now open the possibility to examine this interesting problem.
Our initial spectroscopic characterisation of the Fe/S clusters in Nar1p indicates that there are two different, adjacent Fe/S clusters. One of the clusters gave rise to an unusual rhombic EPR signal, and the other could be observed only indirectly. We speculate that the unusual cluster could be derived from a special cubane and may have additional metal ions. Likely, it is coordinated by the four conserved cysteine residues in the C-terminal part of Nar1p. Modelling of this region of Nar1p corresponding to the active centre H-cluster of iron-only hydrogenases indicated that only one side of the protein pocket containing the binuclear part of the H-cluster is conserved in Nar1p, whereas the other side shows considerable amino-acid exchanges (Nicolet et al, 2002). The nonconserved side therefore may perform the specific function of Nar1p-like proteins. Further spectroscopic, structural and mutational studies are expected to shed light on the exact nature of the Fe/S clusters and to elucidate the mode of action in Fe/S protein maturation. Assembly of the Fe/S clusters on Nar1p in vivo depends on the mitochondrial ISC machinery. Nar1p thus behaves like known [4Fe–4S]-containing proteins of the yeast cytosol, even though Nar1p seems to carry unusual Fe/S clusters. The biosynthesis pathway of Fe/S clusters in bacterial iron-only hydrogenases has not been elucidated yet (Vignais et al, 2001), but based on our experiments we presume that the bacterial ISC machinery is a good candidate for Fe/S cluster assembly. It will be interesting to analyse whether further, specialised components will be required for synthesis of both the H-cluster in hydrogenases and the unusual Fe/S cluster on Nar1p and learn whether these components are related.
Our study provides the first account on the assembly of a nuclear Fe/S protein, the N-glycosylase Ntg2p. It appears that this protein behaves similarly to cytosolic Fe/S apoproteins in that Fe/S cluster association depends on Nar1p. At present, it is unclear where the assembly of Ntg2p occurs, since a fraction of Nar1p may be located in the nucleus. Even though Nar1p does not contain a canonical nuclear targeting sequence, a conspicuous lysine- and arginine-rich motif (RKRX5RKRR; Figure 1) may direct its nuclear targeting. It has to be mentioned though that this sequence is not conserved in other members of the Nar1 protein family, in particular not in the nucleus-located human Narf, which uses another nuclear localisation signal (Barton and Worman, 1999). It will be interesting to study whether Ntg2p may get its Fe/S cluster in the nucleus or already in the cytosol, either co- or post-translationally.
A particularly intriguing aspect of our findings is how, during evolution, Nar1p developed from its ancestor, a bacterial iron-only hydrogenase, to a factor supporting cytosolic Fe/S protein assembly. The eukaryotic cytosol is believed to be derived from the ancestral archaebacterial cell that incorporated an α-proteobacterium as an endosymbiont to eventually lead to a eukaryotic cell containing a mitochondrion. Hence, the archaebacterial cytosol may be expected to be the source of components supporting cytosolic Fe/S protein biogenesis. However, so far, no iron-only hydrogenases have been identified in Archaea (Vignais et al, 2001). It therefore appears that the Nar1p precursor may have escaped the original endosymbiont to gain a new function in the eukaryotic cytosol. In contrast, the P-loop ATPase Cfd1p required for cytosolic Fe/S protein maturation may well have been derived from Archaea, as the homologous Mrp proteins are well conserved in this kingdom (Leipe et al, 2002).
The Nar1 protein family has already caught the attention of bioinorganic chemists, evolutionists and hydrogenase specialists. Our identification of a functional role for a member of this family, yeast Nar1p, will now open the way for detailed biochemical analysis of its molecular mode of action. Future insights into the structure of the Fe/S cofactors, the evolutionary origin and the biochemical function of Nar1p are expected to further enhance our understanding of these fascinating proteins.
Materials and methods
Yeast strains, cell growth and plasmids
The following strains of S. cerevisiae were used: PSY581 (_MAT_α, ura3-52, leu2Δ1, his3Δ200) for localisation experiments. For all other experiments, strain W303-1A (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3112) served as wild type. The Gal-NFS1, Gal-YAH1 and Gal-ATM1 strains were described previously (Kispal et al, 1999; Lange et al, 2000). Mutant strain Gal-NAR1 was derived from W303-1A by exchange of the endogenous NAR1 promoter (nucleotides −197 to −1) for the galactose-inducible GAL1-10 promoter by homologous recombination using a PCR product containing the HIS3 marker gene (Mühlenhoff et al, 2002). Correct insertion of the DNA into the yeast genome was verified by PCR. Cells were grown in rich (YP) and minimal (SC) media, or in minimal medium lacking added iron chloride (‘iron-poor'), each containing the required carbon sources at a concentration of 2% (w/v) unless indicated otherwise (Sherman, 1991). The following yeast plasmids were used: pRS416 containing the MET25 promoter, and pRS426 with the TDH3 promoter (Mumberg et al, 1995). All constructs were verified by DNA sequencing.
Recombinant protein techniques
The NAR1 gene was isolated from genomic DNA of yeast strain W303-1A using PCR and cloned into the appropriate vectors: pET15b (Novagen) for overexpression in E. coli of NAR1 with an N-terminal, hexahistidinyl-tag (for antibody production); pET3a (Novagen) for NAR1 overexpression with a C-terminal _Strep_-tag II encoded in the 3′PCR primer. His-tagged and _Strep_-tagged Nar1p were purified under anaerobic conditions using Ni-NTA agarose (Qiagen, Düsseldorf) or _Strep_-Tactin columns (IBA, Göttingen, Germany), respectively, according to the manufacturer's manuals. Identity and mass of the His-tagged Nar1p were confirmed by MALDI-TOF.
Electron paramagnetic resonance
Purified, de-salted hexahistidinyl-tagged or _Strep_-tagged Nar1p were used for EPR spectroscopy and yielded similar spectra. Low-temperature X-band EPR spectra were recorded with a Bruker ESP 300E cw spectrometer, equipped with a helium flow cryostat ESR910 (Oxford Instruments; Pierik et al, 1992).
Miscellaneous methods
The following published methods were used: manipulation of DNA and PCR (Sambrook and Russell, 2001); transformation of yeast cells (Gietz et al, 1992); enzyme activities of isopropylmalate isomerase, aconitase and succinate dehydrogenase (Kispal et al, 1999; Mühlenhoff et al, 2002); raising of antisera (Harlow and Lane, 1998); in situ immunofluorescence (Krebber et al, 1999); quantitative analysis of iron and sulphur in purified Nar1p (Pierik et al, 1992); quantitation of iron in mitochondria by the colorimetric chelator ferene (Hennessy et al, 1984). Highly specific anti-Nar1p antibodies were affinity-purified from the polyclonal antiserum using nitrocellulose-bound recombinant Nar1p. Radiolabelling and cell lysis of the Fe/S reporter proteins of interest were as described previously (Kispal et al, 1999; Mühlenhoff et al, 2003). All experiments were repeated at least three times. Error bars represent the standard error of the mean value.
Acknowledgments
We thank Drs W Buckel and RK Thauer for generously supporting the EPR spectroscopical experiments, Dr H Worman for providing the NARF and HPRN genes, Dr H Krebber for yeast strain PSY581, Dr H Ulrich for affinity-purified anti-Pol30p antibodies and Dr J Nyalwidhe for MALDI-TOF analysis. Our work was supported by grants of Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 593, European Commission (QLG1-CT-2001-00966), Deutsches Humangenomprojekt and Fonds der Chemischen Industrie.
References
- Adams MW (1987) The mechanisms of H2 activation and CO binding by hydrogenase I and hydrogenase II of Clostridium pasteurianum. J Biol Chem 262: 15054–15061 [PubMed] [Google Scholar]
- Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol Cell Biol 19: 3779–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris JP, Blobel G (1991) Isolation of yeast nuclei. Methods Enzymol 194: 735–749 [DOI] [PubMed] [Google Scholar]
- Barton RM, Worman HJ (1999) Prenylated prelamin A interacts with Narf, a novel nuclear protein. J Biol Chem 274: 30008–30018 [DOI] [PubMed] [Google Scholar]
- Beinert H, Albracht SPJ (1982) New insights, ideas and unanswered questions concerning iron–sulfur clusters in mitochondria. Biochim Biophys Acta 683: 245–277 [DOI] [PubMed] [Google Scholar]
- Bekri S, Kispal G, Lange H, Fitzsimons E, Tolmie J, Lill R, Bishop DF (2000) Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia (XLSA/A) with disruption of cytosolic iron–sulfur protein maturation. Blood 96: 3256–3264 [PubMed] [Google Scholar]
- Cazzola M, Invernizzi R, Bergamaschi G, Levi S, Corsi B, Travaglino E, Rolandi V, Biasiotto G, Drysdale J, Arosio P (2003) Mitochondrial ferritin expression in erythroid cells from patients with sideroblastic anemia. Blood 101: 1996–2000 [DOI] [PubMed] [Google Scholar]
- Chen OS, Hemenway S, Kaplan J (2002) Inhibition of Fe–S cluster biosynthesis decreases mitochondrial iron export: evidence that Yfh1p affects Fe–S cluster synthesis. Proc Natl Acad Sci USA 99: 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2002) A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cell Mol Life Sci 59: 1658–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Talibi D (2001) Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J Biol Chem 276: 7762–7768 [DOI] [PubMed] [Google Scholar]
- Frazzon J, Dean DR (2003) Formation of iron–sulfur clusters in bacteria—an emerging field in bioinorganic chemistry. Curr Opin Chem Biol 7: 166–173 [DOI] [PubMed] [Google Scholar]
- Gerber J, Lill R (2002) Biogenesis of iron–sulfur proteins in eukaryotes: components, mechanism and pathology. Mitochondrion 2: 71–86 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gietz D, StJean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen WR, Albracht SPJ (1982) Analysis of strain-induced EPR-line shapes and anisotropic spin-lattice relaxation in a [2Fe–2S] ferredoxin. Biochim Biophys Acta 702: 61–71 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1998) Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Hennessy DJ, Reid GR, Smith FE, Thompson SL (1984) Ferene—a new spectrophotometric reagent for iron. Can J Chem 62: 721–724 [Google Scholar]
- Horner DS, Heil B, Happe T, Embley TM (2002) Iron hydrogenases—ancient enzymes in modern eukaryotes. Trends Biochem Sci 27: 148–153 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Guiard B, Lill R (1997) The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett 418: 346–350 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are required for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber H, Taura T, Lee MS, Silver PA (1999) Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev 13: 1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Kispal G, Kaut A, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of intra- and extra-mitochondrial Fe/S proteins. Proc Natl Acad Sci USA 97: 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Lisowsky T, Gerber J, Mühlenhoff U, Kispal G, Lill R (2001) An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep 2: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L (2002) Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317: 41–72 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe/S proteins: the essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Mathews R, Charlton S, Sands RH, Palmer G (1974) On the nature of the spin coupling between the iron–sulfur clusters in the eight-iron ferredoxins. J Biol Chem 249: 4326–4328 [PubMed] [Google Scholar]
- Mühlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R (2002) The yeast frataxin homologue Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet 11: 2025–2036 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Nicolet Y, Cavazza C, Fontecilla-Camps JC (2002) Fe-only hydrogenases: structure, function and evolution. J Inorg Biochem 91: 1–8 [DOI] [PubMed] [Google Scholar]
- Nicolet Y, Lemon BJ, Fontecilla-Camps JC, Peters JW (2000) A novel FeS cluster in Fe-only hydrogenases. Trends Biochem Sci 25: 138–143 [DOI] [PubMed] [Google Scholar]
- Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC (1999) Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Struct Fold Des 7: 13–23 [DOI] [PubMed] [Google Scholar]
- Orme-Johnson WH, Orme-Johnson AR (1982) Iron–sulfur proteins: the problem of determining cluster type. In Iron–Sulfur Proteins, Spiro TG (ed) pp 67–95. New York: Wiley [Google Scholar]
- Peters JW (1999) Structure and mechanism of iron-only hydrogenases. Curr Opin Struct Biol 9: 670–676 [DOI] [PubMed] [Google Scholar]
- Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC (1998) X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom resolution. Science 282: 1853–1858 [DOI] [PubMed] [Google Scholar]
- Pierik AJ, Hagen WR, Redeker JS, Wolbert RB, Boersma M, Verhagen MF, Grande HJ, Veeger C, Mutsaers PH, Sands RH et al. (1992) Redox properties of the iron–sulfur clusters in activated Fe-hydrogenase from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem 209: 63–72 [DOI] [PubMed] [Google Scholar]
- Robinson AC, Dean DR, Burgess BK (1987) Iron–molybdenum cofactor biosynthesis in Azotobacter vinelandii requires the iron protein of nitrogenase. J Biol Chem 262: 14327–14332 [PubMed] [Google Scholar]
- Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE (2003) A novel eukaryotic factor for cytosolic Fe–S cluster assembly. EMBO J 22: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR (2001) A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci USA 98: 14322–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G (2002) Maturation of cytosolic iron–sulfur proteins requires glutathione. J Biol Chem 277: 26944–26949 [DOI] [PubMed] [Google Scholar]
- Skovran E, Downs DM (2003) Lack of the ApbC or ApbE protein results in a defect in Fe–S cluster metabolism in Salmonella enterica serovar Typhimurium. J Bacteriol 185: 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BE, O'Donnell MJ, Lang G, Spartalian K (1980) A Mössbauer spectroscopic investigation of the redox behaviour of the molybdenum–iron protein from Klebsiella pneumoniae nitrogenase. Biochem J 191: 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamer W, Cirpus I, Hans M, Pierik AJ, Selmer T, Bill E, Linder D, Buckel W (2003) A two [4Fe–4S]-cluster-containing ferredoxin as an alternative electron donor for 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Arch Microbiol 179: 197–204 [DOI] [PubMed] [Google Scholar]
- Vignais PM, Billoud B, Meyer J (2001) Classification and phylogeny of hydrogenases. FEMS Microbiol Rev 25: 455–501 [DOI] [PubMed] [Google Scholar]