The IL-6- and lipopolysaccharide-induced transcription of hepcidin in HFE-, transferrin receptor 2-, and β2-microglobulin-deficient hepatocytes (original) (raw)
Abstract
The antimicrobial peptide hepcidin appears to play a central role in the regulation of iron homeostasis. In intact animals, iron overload or the injection of lipopolysaccharide (LPS) stimulates transcription of HAMP, the gene that encodes hepcidin. In isolated hepatocytes, IL-6, an inflammatory cytokine the production of which is stimulated by LPS, up-regulates transcription of hepcidin. In contrast, iron has no stimulatory effect on hepcidin expression in isolated hepatocytes. There is apparently a signaling pathway, activated by iron, that is present in the intact animal but not in isolated hepatocytes. Studies in humans and mice have shown that this iron-dependent pathway requires the presence of Hfe, hemojuvelin, and probably transferrin receptor 2 (tfr-2). To determine whether activation of hepcidin transcription by IL-6 also requires Hfe and tfr-2, we have studied mice homozygous for targeted disruption of HFE, β2-microglobulin, and for a truncating mutation of TFR-2. We show that these mutant mice react normally to injection of endotoxin and that their isolated hepatocytes react normally to IL-6. This indicates that the signaling pathway activated by IL-6 does not require either Hfe or tfr-2. Mice with disruption of the gene encoding IL-6 seem to have a blunted response to LPS, but the statistical significance of the small response documented is borderline. It is therefore not clear whether LPS stimulates secretion of cytokines other than IL-6 that may stimulate hepcidin transcription.
The antimicrobial peptide hepcidin, encoded by the HAMP gene, has emerged as a major regulator of iron homeostasis in mice and humans. Transcription of hepcidin mRNA is stimulated by iron-loading hepatocytes of intact mice (1). However, isolated human (2) and murine (1) hepatocytes exposed to iron do not increase transcription of the HAMP gene. Accordingly, there must be a signaling pathway to the HAMP gene that is activated by iron, but that is not fully represented in the isolated cells. In humans (3) and mice (4), up-regulation of HAMP is impaired when there is a deficiency of HFE. A deficiency of hemojuvelin, the product of the recently cloned HJV gene, also seems to prevent the up-regulation of hepcidin. This suggests that Hfe and hemojuvelin play a role in transducing an ironinduced signal that stimulates HAMP transcription. Another protein that may be involved in signal transduction is transferrin receptor 2 (tfr-2) (5), because mutations of the gene encoding this receptor are associated in humans and mice with a phenotype very similar to that of HFE-deficient humans (6) and mice (7). Apart from the evidence that suggests that Hfe, hemojuvelin, and tfr-2 may be involved, nothing is known of the manner in which increased iron stores signal the up-regulation of HAMP transcription, but it has been suggested that cell–cell interaction between macrophages and hepatocytes may play a role (2, 8).
Hepcidin also functions as an antimicrobial peptide, and it is not only iron loading that stimulates its production but also lipopolysaccharide (LPS) (1), which appears to function through the mediation of the inflammatory cytokine IL-6 (8). Although the up-regulation of hepcidin by iron requires the action of HFE and of hemojuvelin, it is not known whether IL-6 stimulation follows the same pathway. To answer this question, we have examined the putative role of Hfe and tfr-2 in the pathway by which IL-6 stimulates hepcidin production in mutant Hfe- and tfr-2-deficient mice and primary hepatocytes derived from such animals. We show that stimulation of the formation of hepcidin transcripts in murine hepatocytes is unimpaired in Hfe or tfr-2-deficient animals. Moreover, mice in which the gene encoding IL-6 has undergone targeted disruption demonstrate that stimulation of hepcidin transcription by LPS may not depend solely on IL-6.
Materials and Methods
Mice. C57BL/6 HFE-/- mice were generously provided by William Sly (Saint Louis University, St. Louis) and backcrossed 10 generations into 129 mice. AKR/J TfR2Y245X mice were generously provided by Robert Fleming (Saint Louis University). B6.129S2-Il6tm1kopf (002650), B10.129P2(B6)-B2mtm1Unc (002454), AKR/J (000648), C57BL/6J (000664), and 129 × 1/SvJ (000691) mice were obtained from The Jackson Laboratory. These animals were maintained in our animal quarters and backcrossed against their congenic strains.
LPS Treatment. Mice were fasted overnight and then treated with LPS (Westphal preparation from Escherichia coli 055:B5, Sigma) (1 μg/g i.p.) or an equivalent volume of saline. Livers were removed 6 h posttreatment and total RNA isolated using RNeasy mini kit (Qiagen, Valencia, CA). This is the time at which maximal response of hepcidin transcription has been noted (9).
Hepatocyte Isolation and IL-6 Treatment. Mouse hepatocytes were isolated after liver perfusion at a rate of 5 ml/min with buffers warmed to 44°C, unless otherwise indicated. Each liver was perfused with (i) 25 ml of 0.5 mM EGTA in PBS, (ii) 10 ml of PBS, (iii) 20 ml of liver digest media (GIBCO/BRL Life Technologies) supplemented with an additional 0.15 mg/ml collagenase (hepatocyte grade, GIBCO/BRL Life Technologies), this final step at a rate of 2 ml/min. The livers were removed and transferred to a chilled Petri dish containing 15 ml of chilled liver wash media (GIBCO/BRL Life Technologies). Dissociated cells were isolated by gentle rubbing of the liver with a rubber policeman. Hepatocytes were pelleted at 100 × g for 4 min at 4°C and washed once with liver wash media. The washed hepatocytes were then resuspended in 25 ml of Williams incomplete media (GIBCO/BRL Life Technologies) and layered onto a 50-ml conical tube containing 20 ml of Percoll (Sigma) (1 part 10× HBSS and 9 parts Percoll). The hepatocytes were mixed with Percoll by inverting five times. Hepatocytes were pelleted by centrifugation at 300 × g for 10 min at 4°C. The hepatocytes were then washed with 12 ml of Williams media, pelleting at 100 × g for 4 min. The cells were resuspended in 6 ml of complete Williams incomplete media with 5% FBS and seeded onto collagen-coated Biocoat six-well plates (BD Biosciences) at a density of 2.4 × 105 cells per well. After 22 h, the media were changed, and the cells were treated with IL-6 at 20 ng/ml (R & D Systems). RNA was isolated by using RNeasy (Qiagen), and RT-PCR was performed.
RT-PCR. cDNA was synthesized from up to 10 μg of RNA in a 50-μl reaction mixture containing 1.5 μg of oligo dT15, 1× reverse transcriptase buffer, 10 mM DTT, 1 mM dNTPs, 1 unit of Rnasin, and 1 unit of Moloney's murine leukemia virus reverse transcriptase (Invitrogen Life Technologies) for 1 h at 37°C.
Real-Time PCR. Real-time PCR was carried out on a Bio-Rad (Hercules, CA) iCycler. Primers were designed with the help of Beacon Designer (Bio-Rad). Primers were designed for analysis of murine hepcidin and murine transthyretin (TTR) as a hepatic specific control.
The hepcidin primers, which amplify both mouse hepcidin 1 and 2, were 5′-TGTCTCCTGCTTCTCCTCCT-3′ and 5′-CTCTGTAGTCTGTCTCATCTGTTG-3′. The hepcidin 1 and 2 FAM-labeled probe was 5′-/56-FAM/CCAGCCTGAGCAGCACCACCTATC/3BHQ_1/-3′. The amplification primers for TTR were 5′-TTTCACAGCCAACGACTCTGG-3′ and 5′-GGCAAGATCCTGGTCCTCCT-3′, and the labeled probe was 5′-/56-FAM/TCGCCACTACACCATCGCAGCCCT/3BHQ_1/-3′.
The iCycler amplification system was iQ Supermix (Bio-Rad) with the addition of MgCl2 to a final concentration of 4.6 mM. Three microliters of synthesized cDNA or standards was used in a 25-μl amplification system. All samples were assayed in duplicate or triplicate. After 40 amplification cycles, threshold cycle values were automatically calculated, and attomol of starting cDNA were calculated from a standard curve covering a range of four orders of magnitude. The hepcidin standard curve ranged from 0.09 to 900 attomol per 25-μl reaction, and the TTR standard curve ranged from 0.036 to 360 attomol per reaction. Ratios of hepcidin to TTR starting quantity were calculated. Experimental samples were expressed as a percentage of the concurrent control.
Results
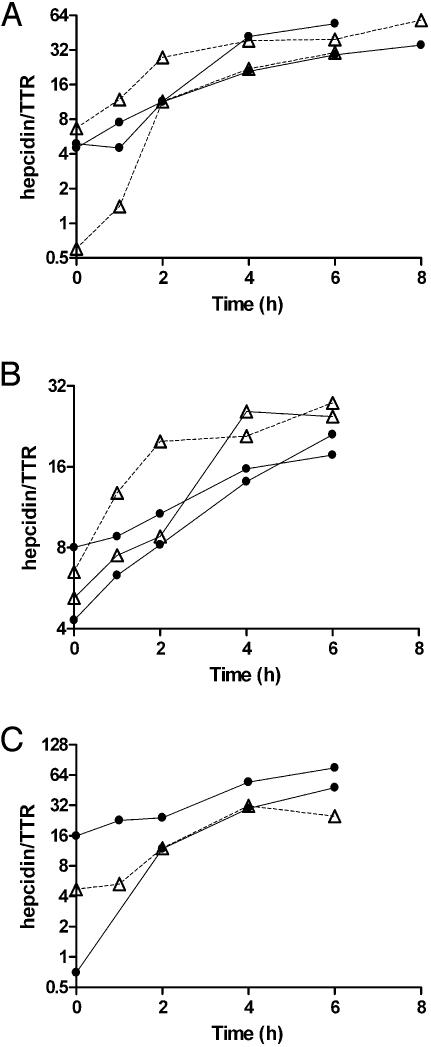
Studies in Primary Hepatocyte Cultures. Isolated hepatocytes from HFE-/-, TfR2 Y245X, β2-microglobulin-/-, and normal mice from their background strains were treated with IL-6, and the expression of hepcidin was measured at 1, 2, 4, and 6 h. The induction of hepcidin expression in the hepatocytes from the mutant strains paralleled the response from the control congenic strains (Fig. 1).
Fig. 1.
The response of isolated hepatocytes to treatment with IL-6. (A) The response of hepatocytes from HFE knockout mice. (B) tfr-2 mutant mice. (C)A β-microglobulin knockout mouse. In each, the response to IL-6 is compared with that of hepatocytes from the congenic strain also treated with IL-6. Open triangles represent the deficient mice; filled circles are the controls.
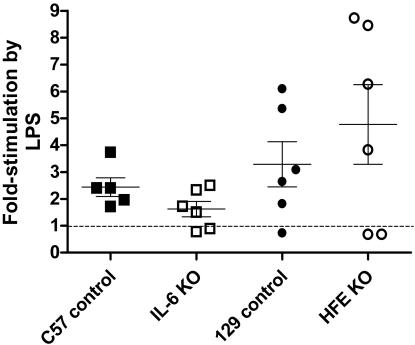
Studies in Mice Injected with LPS. Control, HFE-/-, and TfR2 Y245X mutant mice were examined for the induction of liver hepcidin expression in response to LPS treatment. Control, HFE-/-, and TfR2 Y245X mice were all responsive to LPS treatment, but there was marked variability between the responses manifested by different animals with identical genetic backgrounds (Fig. 2 and data not shown).
Fig. 2.
The hepcidin mRNA response of mice 6 h after i.p. injection of 1 μg of LPS per gram of body weight. Responses represent the ratio of hepcidin cDNA to TTR cDNA as measured by real-time PCR (see Materials and Methods). The results are presented as the percentage of the average hepcidin/TTR ratio documented in mice of the same age and strain that were injected with saline and killed after 6 h. Each point represents one mouse. The dashed line indicates the level at which there is no stimulation of hepcidin transcription by LPS. The mean and one standard error of the mean are shown.
To determine whether the hepcidin response to LPS is mediated wholly or partly through IL-6, IL-6-/- mice were treated with LPS and the liver hepcidin expression examined. Hepcidin transcription seemed to be mildly stimulated by LPS in IL-6-/- mice (P = 0.05) but significantly less so than the induction by LPS in the congenic strain (P < 0.05) (Fig. 2).
Discussion
Previously published studies indicate that hepcidin expression is up-regulated by iron loading of mice and by LPS treatment of mice (1). In vitro, iron fails to up-regulate hepcidin transcription in hepatocytes from normal animals (1, 2). One may therefore conclude that there is a signaling pathway activated by iron that stimulates the transcription of hepcidin, and that this pathway is not fully represented in isolated hepatocytes. Little is known of how this iron-dependent pathway functions, but the requirement for Hfe and hemojuvelin in the transduction of the iron signal to hepcidin transcription can be inferred from the fact that HFE- and hemojuvelin-deficient patients do not manifest increased hepcidin production when they become iron-loaded (2, 10). The same is very likely true in the case of trf-2, a deficiency of which has the same phenotype as HFE deficiency.
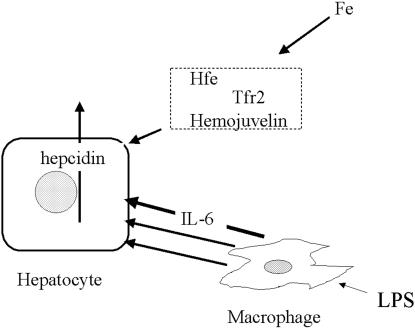
LPS also activates the transcription of HAMP. Monocyte-conditioned media stimulate hepatocytes hepcidin transcription, and this stimulation is not abrogated by anti-IL-1 (2). Our studies were designed chiefly to answer the question of whether the signaling pathway initiated by LPS, functioning presumably through IL-6, depended, like the iron-initiated pathway, on the presence of Hfe and trf-2. The study of primary hepatocytes cultures from HFE knockout and TFR-2 mutant mice shows clearly that these cells are readily stimulated to produce hepcidin message by IL-6. Thus this pathway requires neither Hfe nor trf-2 to function. To the extent that there may be common steps in the stimulation of hepcidin production with iron on the one hand and IL-6 on the other, it is clear that the IL-6-activated pathway is independent of the Hfe and presumably from the trf-2 required by the iron pathway or merges with it downstream of Hfe and trf-2 (Fig. 3).
Fig. 3.
Schematic representation of the regulation of transcription of hepcidin by the iron pathway (above) and inflammatory (LPS/IL-6 pathway) (below). The iron pathway requires Hfe, hemojuvelin, and probably tfr-2 outside of the hepatocytes. These components are not required for the inflammatory pathway. Although IL-6 is an effective mediator of the response to LPS, some other mediators formed in response to LPS may also stimulate the transcription of hepcidin, although the evidence for this is unclear.
In this respect, the results of our studies differ from those recently reported by Roy et al. (11), namely, that Hfe-deficient mice did not respond to LPS stimulation and that they failed to lower their serum iron level in a normal fashion. In our studies, we found, as have others, that the response to LPS differs strikingly from animal to animal. In our studies, two of six _HFE_-/- mice did have a suboptimal hepcidin response to LPS stimulation, but the other four had a robust unmistakable response. In fact, the average response of the _HFE-_deficient mice was slightly greater than that of congenic controls. The dose and timing of LPS injection differed from that used by Roy et al. (11). The differences between our studies and those previously reported may also be due to the marked variability of studies carried out in intact animals. However, the results of our studies are supported by the more robust measurements that we have been able to make in primary hepatocytes. Roy et al. (11) also found that Hfe-deficient animals failed to appropriately lower their serum iron levels 24 h after LPS injection, a finding that might be considered to be due to an inadequate hepcidin response. But the failure of these animals to lower their serum iron in response to LPS injection could well have been due to the fact that these animals, in contrast to the controls, were iron loaded.
Because the response of IL-6-deficient mice to LPS injection seemed to be blunted but not abrogated, it may be that stimulation by LPS is due not only to the production of IL-6 but also possibly to the production of other cytokines. Further studies will be needed to determine whether there are other LPS-induced mediators that stimulate the production of hepcidin.
Acknowledgments
This work was supported by National Institutes of Health Grants DK53505 and RR00833 and the Stein Endowment Fund.
Abbreviations: LPS, lipopolysaccharide; tfr-2, transferrin receptor 2; TTR, murine transthyretin.
References
- 1.Pigeon, C., Ilyin, G., Courselaud, B., Leroyer, P., Turlin, B., Brissot, P. & Loreal, O. (2001) J. Biol. Chem. 276**,** 7811-7819. [DOI] [PubMed] [Google Scholar]
- 2.Nemeth, E., Valore, E. V., Territo, M., Schiller, G., Lichtenstein, A. & Ganz, T. (2003) Blood 101**,** 2461-2463. [DOI] [PubMed] [Google Scholar]
- 3.Gehrke, S. G., Kulaksiz, H., Herrmann, T., Riedel, H. D., Bents, K., Veltkamp, C. & Stremmel, W. (2003) Blood 102**,** 371-376. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad, K. A., Ahmann, J. R., Migas, M. C., Waheed, A., Britton, R. S., Bacon, B. R., Sly, W. S. & Fleming, R. E. (2002) Blood Cells Mol. Dis. 29**,** 361-366. [DOI] [PubMed] [Google Scholar]
- 5.Frazer, D. M. & Anderson, G. J. (2003) Blood Cells Mol. Dis. 30**,** 288-297. [DOI] [PubMed] [Google Scholar]
- 6.Camaschella, C., Roetto, A., Cali, A., De Gobbi, M., Garozzo, G., Carella, M., Majorano, N., Totaro, A. & Gasparini, P. (2000) Nat. Genet. 25**,** 14-15. [DOI] [PubMed] [Google Scholar]
- 7.Fleming, R. E., Ahmann, J. R., Migas, M. C., Waheed, A., Koeffler, H. P., Kawabata, H., Britton, R. S., Bacon, B. R. & Sly, W. S. (2002) Proc. Natl. Acad. Sci. USA 99**,** 10653-10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganz, T. (2003) Blood 102**,** 783-788. [DOI] [PubMed] [Google Scholar]
- 9.Yeh, K. Y., Yeh, M. & Glass, J. (2004) Am. J. Physiol. 286**,** G385-G394. [DOI] [PubMed] [Google Scholar]
- 10.Papanikolaou, G., Samuels, M. E., Ludwig, E. H., MacDonald, M. L., Franchini, P. L., Dube, M. P., Andres, L., MacFarlane, J., Sakellaropoulos, N., Politou, M., et al. (2004) Nat. Genet. 36**,** 77-82. [DOI] [PubMed] [Google Scholar]
- 11.Roy, C. N., Custodio, A. O., De Graaf, J., Schneider, S., Akpan, I., Montross, L. K., Sanchez, M., Gaudino, A., Hentze, M. W., Andrews, N. C., et al. (2004) Nat. Genet. 36**,** 481-485. [DOI] [PubMed] [Google Scholar]