Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity (original) (raw)
. Author manuscript; available in PMC: 2015 May 12.
Summary
Bacterial cytokinesis is driven by the septal ring apparatus, the assembly of which in Escherichia coli is directed to mid-cell by the Min system. Despite suffering aberrant divisions at the poles, cells lacking the minCDE operon (Min–) have an almost normal growth rate. We developed a generally applicable screening method for synthetic lethality in E. coli, and used it to select for transposon mutations (slm) that are synthetically lethal (or sick) in combination with Δ_min-CDE_. One of the slm insertions mapped to envC (yibP), proposed to encode a lysostaphin-like, metallo-endopeptidase that is exported to the periplasm by the general secretory (Sec) pathway. Min– EnvC– cells showed a severe division defect, supporting a role for EnvC in septal ring function. Accordingly, we show that an EnvC–green fluorescent protein fusion, when directed to the periplasm via the twin-arginine export system, is both functional and part of the septal ring apparatus. Using an in-gel assay, we also present evidence that EnvC possesses murein hydrolytic activity. Our results suggest that EnvC plays a direct role in septal murein cleavage to allow outer membrane constriction and daughter cell separation. By uncovering genetic interactions, the synthetic lethal screen described here provides an attractive new tool for studying gene function in E. coli.
Introduction
Cell division in Escherichia coli is a complex process involving the co-ordinated invagination of the three cell envelope layers to form two new daughter cell poles. The process requires the activities of, at least, 10 essential division proteins, which are either cytoplasmic (FtsA and FtsZ) or inner membrane (FtsB, FtsI, FtsK, FtsL, FtsN, FtsQ, FtsW and ZipA) species. All 10 proteins localize to a membrane-associated ring structure that is thought to constitute a molecular machine capable of promoting division (Errington et al., 2003).
The initial step in septal ring assembly appears to be the polymerization of the tubulin-like FtsZ protein into a ring (the Z-ring) just underneath the cytoplasmic membrane (Bi and Lutkenhaus, 1991; Errington et al., 2003). Z-ring formation requires at least one of the known FtsZ-binding proteins, FtsA or ZipA, which incorporate into the ring independently of each other (Hale and de Boer, 1999; Liu et al., 1999; Pichoff and Lutkenhaus, 2002). After the assembly of FtsZ, FtsA and ZipA, the remaining components join the ring in mostly linearly dependent fashion: FtsK, FtsQ, FtsB + FtsL, FtsW, FtsI and, finally, FtsN (Chen and Beckwith, 2001; Buddelmeijer et al., 2002; Hale and de Boer, 2002).
In addition to the essential division proteins, two non-essential septal ring components, ZapA (YgfE) and AmiC, were identified recently (Heidrich et al., 2001; Gueiros-Filho and Losick, 2002; Bernhardt and de Boer, 2003; Johnson et al., 2004). ZapA is a cytoplasmic FtsZ-binding protein originally discovered in Bacillus subtilis. By analogy with the other FtsZ-binding proteins, FtsA and ZipA, ZapA is likely to join FtsZ polymers at a very early stage in septal ring assembly (Gueiros-Filho and Losick, 2002). AmiC is a periplasmic amidase that cleaves the septal murein and promotes daughter cell separation (Heidrich et al., 2001). AmiC requires FtsN for recruitment to the septal ring and, unlike all other known ring components, does not appear to localize until about the time at which constriction is initiated (Bernhardt and de Boer, 2003). A number of additional septal ring components are likely to exist. Important challenges are to identify the full complement of septal ring components and to determine how they interact to form a functional division apparatus.
Z-ring assembly is directed to mid-cell by two partially redundant negative regulatory systems, nucleoid occlusion and the Min system (Errington et al., 2003). By an as yet unknown mechanism, Z-ring assembly is inhibited in the close vicinity of nucleoids such that assembly is limited to the polar regions and the space between segregated nucleoids (Mulder and Woldringh, 1989; Sun et al., 1998; Yu and Margolin, 1999). The Min system blocks polar divisions and can direct Z-ring formation to mid-cell even in the absence of nucleoids (de Boer et al., 1989; Sun et al., 1998). In E. coli, this system is composed of three proteins, MinC, MinD and MinE, encoded by the minB operon (de Boer et al., 1989). MinC and MinD form a division inhibitor complex that rapidly oscillates from one cell pole to the other (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a,b; Shih et al., 2003), in a process that is driven by interactions of MinD with ATP, itself, the membrane and MinE (Hu and Lutkenhaus, 2001; Hu et al., 2002; 2003; Suefuji et al., 2002; Lackner et al., 2003). MinC blocks FtsZ polymerization and Z-ring formation (Hu et al., 1999; Pichoff and Lutkenhaus, 2001; Johnson et al., 2002), and it is the pole-to-pole oscillation of this division inhibitor that is proposed to direct assembly of the division apparatus to mid-cell (Hale et al., 2001; Howard et al., 2001; Meinhardt and de Boer, 2001; Kruse, 2002; Huang et al., 2003).
Despite suffering a significant number of non-productive polar divisions, cells lacking the minCDE operon have a nearly normal growth rate (Åkerlund et al., 1992; Donachie and Begg, 1996). Cell growth is severely affected, however, when D_minCDE_ is combined with the thermosensitive ftsZ_84 allele (Yu and Margolin, 2000), or with a rodA mutation resulting in the loss of rod shape (Corbin et al., 2002). Given these results, we reasoned that a random screen for mutations synthetically lethal (or ‘sick’) with D_minCDE (slm mutations) would be useful for the identification of (novel) factors involved in cell division and shape determination. We therefore developed a synthetic lethal screen for E. coli based on an unstable mini-F vector and used it to isolate transposon mutants with a tight Slm phenotype. This report describes the details of the screen and the characterization of two mutants, _slm_3 (ybeB::EZTnKan-2) and _slm_11 (envC::EZTnKan-2).
Results
The slm screen
The screen for slm mutants was modelled after the classic Saccharomyces cerevisiae synthetic lethal screen (Bender and Pringle, 1991), which relies on a colony-sectoring phenotype to identify mutants that retain a normally unstable plasmid. Strain TB15/pTB8 [Δ_lacIZYA_ Δ_minCDE_/_bla lacI_q Plac::_minCDE lacZ_] is an MG1655 derivative lacking the min and lac operons. It harbours a mini-F vector (pTB8) containing minCDE and lacZ under the control of the lac promoter (Plac). Because the vector lacks the natural F-factor stabilization systems (Koop et al., 1987), it is unstable and easily lost. This was readily apparent when TB15/pTB8 was plated on non-selective LB agar containing IPTG and Xgal (LB-IX agar). About half of the colonies were solid-white, indicating that they derived from a plasmid-free cell, while the other half were blue with a sectored appearance (sectored-blue), indicating rapid loss of the plasmid as colonies developed (Fig. 1).
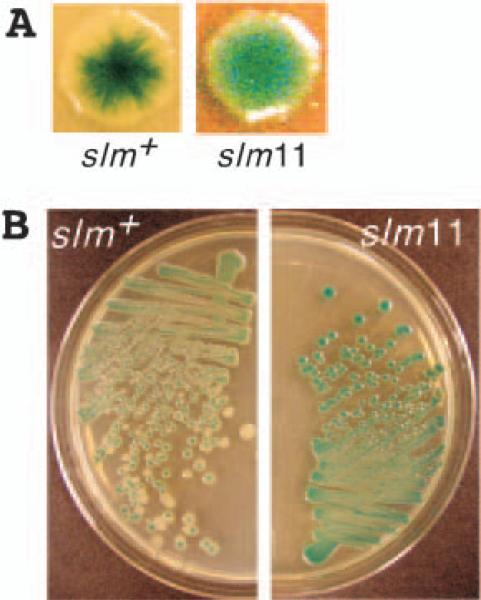
Fig. 1.
Colony sectoring and the screen for slm mutants. Colony phenotypes of strain TB15/pTB8 [Δ_lacIZYA_ Δ_minCDE_/Plac::minCDE _lacZ_] (left) and a _slm_11 derivative (right) grown on non-selective LB agar supplemented with 500 μM IPTG and 60 μg ml−1 Xgal. Note that slm+ cells produced both solid-white and sectored-blue colonies with an asterisk-like appearance, indicating rapid loss of pTB8, whereas _slm_11 colonies were almost exclusively solid-blue, indicating selective pressure to retain the plasmid.
A. Typical sectored (slm+) and solid-blue (slm11) colonies from (B) in more detail. Although not visible here, the slm11 strain also produced some tiny white colonies, consisting of highly filamentous cells.
TB15/pTB8 was mutagenized with the EZTnKan-2 transposome (Epicentre), and the resulting library of about 104 independent transposon mutants was screened for mutants forming solid-blue colonies on LB-IX agar at 30°C. Such mutants presumably cannot lose pTB8 because the EZTn insertion renders the minCDE operon essential. Out of ≈ 30 000 colonies, 62 potential slm mutants were selected. To eliminate false positives, three phenotypic criteria were applied: (i) the mutants should yield solid-blue colonies upon purification on LB-IX agar; (ii) if solid-white colonies were also obtained, they should have a growth defect relative to the solid-blue colonies; and (iii) as minCDE is under Plac control on pTB8, the mutants should be IPTG dependent for growth. Two mutants, _slm_3 and _slm_11, satisfied all three criteria (Figs 1B and 2A). The remaining mutants gave rise to either sectored-blue and solid-white colonies similar to the parental strain or solid-blue and solid-white colonies of similar size.
Fig. 2. Growth and morphology of the slm mutants.
A. Growth phenotypes of TB15/pTB8 [Δ_lacIZYA_ Δ_minCDE_/Plac::_minCDE lacZ_] (left) and its _slm_3 (centre) and _slm_11 (right) derivatives on LB agar with (1) or without (2) 500 μM IPTG. Note the poor growth of the _slm_3 and _slm_11 mutants in the absence of IPTG. B. DIC micrographs of TB15/pTB8 (1 and 2) and its _slm_3 (3 and 4) and _slm_11 (5 and 6) derivatives grown in liquid LB with (1, 3 and 5) or without (2, 4 and 6) 500 μM IPTG. Cells were grown to an OD600 of 0.6–0.7 and fixed. Bar equals 5 μm.
C. Correction of the _slm_3 phenotype by multicopy pbpA rodA. DIC micrographs of strain TB81 [_slm_3 Para::_minCDE_] carrying pMLB1113 [vector] (1), pTB58 [Plac::_ybeBA pbpA rodA_] (2) or pTB59 [Plac::_pbpA rodA_] (3). Cells were grown in LB with no (1 and 2) or 50 μM (3) IPTG to an OD600 of 0.6–0.8 and fixed. Note that cells were Min– because arabinose was omitted from the medium. Bar equals 2 μm.
Both _slm_3 and _slm_11 mutants displayed striking morphological defects (Fig. 2B). When grown in the presence of IPTG (Min+ condition), _slm_3 cells were about 60% wider than the parental slm+ cells (Fig. 2B3). Without IPTG (Min–), the cells became elongated, developed exaggerated shape defects and lysed more frequently than normal (Fig. 2B4; data not shown). _slm_11 cells were slightly filamentous when grown in the presence of IPTG (Fig. 2B5). Many of the cells appeared as short chains or long pairs of about four times the length of a slm+ cell, suggesting a delay in constriction and daughter cell separation (Fig. 2B5). Without IPTG, _slm_11 cells formed very long filaments (Fig. 2B6). For reasons that are unclear, the observed filamentation was more severe on solid relative to liquid media (data not shown).
The slm insertions were mapped by arbitrary polymerase chain reaction (PCR) (O'Toole and Kolter, 1998). The _slm_3 one is located after nucleotide (nt) 5948 of GenBank accession AE000168, disrupting codon 19 of ybeB (Fig. 3A). The _slm_11 insertion is located after nt 4814 of GenBank accession AE000439, just after codon 183 of yibP (Fig. 3B).
Fig. 3.
Maps of plasmids, phage constructs and positions of the transposon insertions. Diagrams of the ybeBA pbpA rodA (A) and envC (B) loci are shown, indicating the location and orientation of the EZTnKan-2 insertions responsible for the Slm phenotype. The SWISSPROT annotations for YbeB (P05848) and EnvC (P37690) were used to construct the figure. Plasmid and phage inserts (solid lines), the lac or phage T7 promoters (arrows), gfp_mut2 (grey box), the FKH tag (white box), the insertion of the AmiC signal sequence coding region (ss_amiC) and the presence of a T7 gene10 ribosome binding site (*) are indicated. The predicted domain organization of EnvC is shown in (B). The signal sequence (SS), predicted coiled-coil region (CC) and M37 metallo-endopeptidase domain (M37) are shown. The relevant amino acid residue numbers for each domain are given above the diagram. The numbers were derived from Hara et al. (2002) but modified to be consistent with the SWISSPROT annotation.
slm3 affects expression of the shape genes pbpA and rodA
ybeB encodes a member of the highly conserved iojap protein family (Bateman et al., 2002). Iojap is implicated in proper chloroplast development in plants, but its precise role remains obscure (Han et al., 1992). Nothing is known about the function(s) of these proteins in bacteria except that the viability of slm3 cells now indicates that ybeB is not essential to E. coli. The gene lies upstream of ybeA, of unknown function, and the shape genes pbpA and rodA (Fig. 3A), which are involved in maintaining the rod shape of the cell (Spratt et al., 1980).
Whether the wide-cell phenotype resulted from inactivation of ybeB itself or a possible polar effect on the expression of downstream genes was tested by complementation analyses. To this end, plasmids pTB69 [Plac::_ybeBA_], pTB58 [Plac::_ybeBA pbpA rodA_] and pTB59 [Plac::_pbpA rodA_] (Fig. 3A) were introduced into strains TB75 [_slm_3] and TB81 [Para::_minCDE slm_3]. TB81 requires arabinose for minCDE expression because the chromosomal min promoter (P1) (de Boer et al., 1989) was replaced with the araBAD promoter (see Supplementary material). Expression of ybeBA from pTB69 did not suppress the shape or the Slm phenotype caused by the _slm_3 allele (not shown). In contrast, both pTB58 and pTB59 could fully correct the shape phenotype of TB75, as well as the shape and Slm phenotypes of TB81 that are seen in the absence of arabinose (Fig. 2C; data not shown). Therefore, the _slm3_-associated phenotypes most probably result from reduced expression of pbpA and/or rodA. As the combination of Min– with RodA– is known to be poorly tolerated (Corbin et al., 2002), the isolation of slm3 bolstered our confidence in the screening method.
Moderate overexpression of ftsQAZ from the low-copy-number plasmid pTB63 suppressed the growth defect of slm3 mutants under Min– conditions without affecting their increased width (data not shown). This result suggests that the levels of division proteins become limiting when _slm_3 cells lose Min function (see Discussion).
The EnvC– (YibP–) and EnvC– Min– phenotypes
The yibP open reading frame (ORF) was shown recently to be allelic with envC (Hara et al., 2002), a division mutant isolated in the early 1970s (Rodolakis et al., 1973). The envC mutants in these earlier studies grew as short chains of elongated cells (Rodolakis et al., 1973; Hara et al., 2002), much like the slm11 cells under Min+ conditions (Fig. 2B5). Electron micrographs of the original envC mutant PM61 by Rodolakis et al. (1973) suggest that this cell separation phenotype results from inefficient splitting of the septal murein, causing a delay in outer membrane (OM) constriction.
The division defects of EnvC– cells become more severe when incubated at high temperature (42°C) in low-salt (<0.5%) medium (Hara et al., 2002; Ichimura et al., 2002; data not shown). Under these conditions, cells form longer and smoother filaments that were reported to contain very few Z-rings (Ichimura et al., 2002). Our slm screen and subsequent experiments were all performed at 30°C.
As cells carrying any of the envC lesions studied so far might still be capable of producing potentially functional EnvC fragments (Rodolakis et al., 1973; Hara et al., 2002; Ichimura et al., 2002), we constructed strains in which the entire envC coding region was replaced with an evictable aph cassette or with the frt scar that remains after eviction of the cassette (Datsenko and Wanner, 2000). Strains with either Δ_envC_ allele displayed the same cell separation defect as the slm11 cells under Min+ conditions (see below; data not shown).
Strains TB35 [Δ_envC_::aph_] and TB58 [Para::minCDE Δ_envC::_aph_] were used to investigate the Slm phenotype of envC mutants in more depth. TB58 recapitulated the Slm phenotype of the original Slm11 strain and formed long filaments when minCDE expression was not induced with arabinose (Fig. 4D). Importantly, plasmid pTB25 [Plac::_envC_] (Fig. 3B) corrected this phenotype, indicating that it was indeed caused by the loss of envC rather than by polar effects on nearby genes (data not shown). Filamentation was not elicited by the SOS response, as a _recA_– derivative of TB58 (TB62) showed the same EnvC– Min– phenotype (data not shown). In addition, as judged by quantitative immunoblotting, FtsZ levels were not affected in EnvC– or EnvC– Min– mutants (data not shown). Interestingly, however, the filamentation phenotype and associated growth defect of TB58 cells lacking a functional Min system were almost completely suppressed upon mild overexpression of ftsQAZ from the low-copy vector pTB63 [_ftsQAZ_] (data not shown).
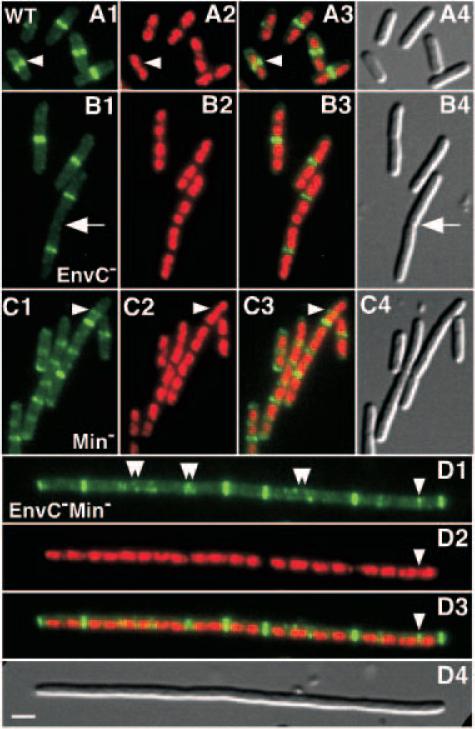
Fig. 4.
Septal ring and nucleoid positioning in EnvC– and EnvC– Min– mutants. Micrographs show representative fields of TB28(λCH151) [wt (Plac::zipA-gfp)] (A), TB35(λCH151) [Δ_envC_::aph (Plac::zipA-gfp)] (B), TB57(λCH151) [Para::minCDE (Plac::zipA-gfp)] (C) and TB58(λCH151) [Δ_envC_::aph Para::minCDE (Plac::zipA-gfp)] (D) cells grown in LB with 50 μM IPTG to an OD600 of 0.6. The cells were fixed, stained with DAPI and viewed for GFP fluorescence (1, pseudocoloured green), DAPI fluorescence (2, pseudocoloured red) and by DIC (4). (3) Overlays of the GFP and DAPI channels. Single arrowheads point to ZipA–GFP rings forming in regions with significant DAPI staining, and double arrowheads point to aberrant ZipA–GFP structures. The arrow in (B) points to a cell constriction without an associated ZipA–GFP ring. Bar equals 2 μm.
Septal ring formation in EnvC– and EnvC– Min– mutants
To understand better the EnvC– phenotype, we compared FtsZ/septal ring and nucleoid distribution patterns in TB28 [wt_] and TB35 [Δ_envC::_aph_] cells containing the prophage λCH151 [Plac::_zipA-gfp_]. Cells were grown in LB media containing 50 μM IPTG, fixed, stained with 4′,6′ diamidino-2-phenylindole (DAPI) and viewed by fluorescence and differential interference contrast (DIC) microscopy. Using these conditions, the majority of wild-type cells showed a single ZipA–green fluorescent protein (GFP) ring at mid-cell, situated in a gap between two DAPI-stained nucleoids (Figs 4A and 5A). Importantly, this level of zipA–gfp expression did not appear to affect cell division, and aberrant structures, such as double rings or spirals, were only rarely observed.
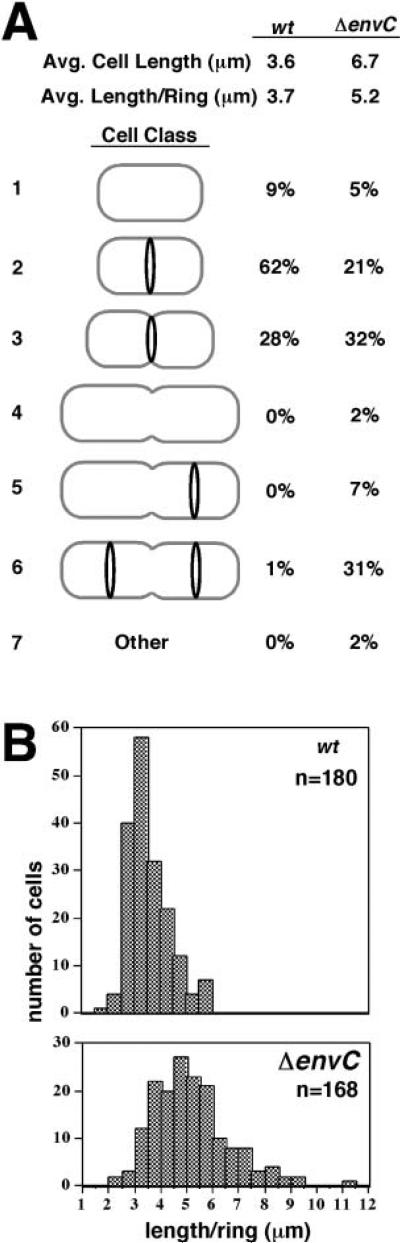
Fig. 5. Measurement of cell length and ZipA–GFP ring formation in EnvC– cells.
A. Cell lengths and ZipA–GFP ring distributions were measured for 197 TB28(λCH151) [wt (Plac::zipA-gfp)] cells and 181 TB35(λCH151) [Δ_envC_::aph (Plac::zipA-gfp)] cells from the experiment described in Fig. 4. The average cell length and length per ZipA–GFP ring (L/R ratio) are given. The types of cells observed were separated into seven classes: (1) unconstricted cells without a ring; (2) unconstricted cells with a medial ring; (3) medially constricting cells with a medial ring; (4) medially constricting cells with no ring; (5) medially constricting cells with one ring at a quarter position; (6) medially constricting cells with a ring at each quarter position; and (7) other types of cells. The ZipA–GFP rings in cells of class 5 tended to be very dim, suggesting that they just began to form. Many cells in class 6 had very shallow constrictions at the quarter positions. The percentage of each cell class observed is indicated to the right of the cartoons. Note that the cartoons do not reflect the relative sizes of the cells in each class. EnvC– cells in classes 3 and 6 were often twice as long as the typical wild-type cells in class 3.
B. Histograms indicating the number of cells with a particular L/R ratio. This ratio was used to normalize for the high frequency of TB35(λCH151) with two rings. The data from (A), excluding the minority of cells without rings, were used to generate the histograms.
Like the original slm_11 mutant, TB35 [Δ_envC::_aph_] cells were elongated, and many were present as pairs (Figs 2B5, 4B and 5A). Ring morphology and nucleoid segregation appeared to be normal in the majority of cells (Fig. 4B). However, the average length to ring (L/R) ratio was significantly larger in EnvC– (5.2 μm) than in wild-type (3.7 μm) cells (Fig. 5A). A comparison of the L/R ratio distributions in ring-containing cells showed a shift to a greater length value (by 1 μm or more) in the EnvC– population, relative to wild type (Fig. 5B). This shift indicates that assembly of a visible ring is significantly delayed in the average mutant cell. The distribution is also broader, suggesting that, once formed, the average ring in EnvC– cells may persist for a longer period of time than normal (Fig. 5B).
Strikingly, a significant number of the long pairs of EnvC– cells, representing 40% of the total population, did not have ZipA–GFP rings associated with their medial constrictions. In the vast majority of these cells, rings were instead present at one or both quarter positions (Figs 4B and 5A). This is in stark contrast to wild-type cells, which almost always have rings present at constrictions (Fig. 5A). The constrictions without associated ZipA–GFP rings might represent failed septa where septal rings disassembled prematurely. If this were the case, however, the cells in classes 5 and 6 of Fig. 5A would be expected to give rise to a significant number of cells with deep constrictions at their quarter positions. Cells of this type were only observed rarely (4/181). An alternative possibility, consistent with the electron micrographs of Rodolakis et al. (1973), is that invagination of the inner membrane (IM) and murein layer was completed at these sites whereas that of the OM was delayed at an early stage. In this case, septal rings are not associated with many of the constrictions because they disassemble upon completion of IM fusion, well before OM invagination is completed. An interesting implication of this phenotype, and probably of chaining phenotypes in general, is that the OM is able to invaginate without an underlying septal ring structure, perhaps using the septal murein as a track to guide its movement.
We next analysed ring and nucleoid distributions in Min– and EnvC– Min– cells. In Min– cells, ZipA–GFP structures were observed at almost all nucleoid-free positions (Fig. 4C). These structures varied markedly in intensity and did not always appear as discrete rings; spirals and double rings were commonly observed (Fig. 4C). This is consistent with previous results on FtsZ patterns in Min– cells (Yu and Margolin, 2000). Similar patterns of ZipA–GFP and nucleoid staining were observed in EnvC– Min– filaments (Fig. 4D). Aberrant ZipA–GFP structures were easier to see in these longer cells (Fig. 4D), but this was not due to the envC lesion per se because they were also observed at a similar frequency in longer Min– cells and in Min– FtsN– filaments (data not shown).
We conclude from these images that the severe division defect of EnvC– Min– cells was not the result of a failure in the early stages of septal ring assembly.
In all strains examined, we occasionally observed ZipA–GFP structures in locations where the nucleoids were not visibly segregated (Fig. 4A, C and D). Similar examples were described in a report using immunofluorescence to stain FtsZ (Yu and Margolin, 2000). Such occurrences suggest that either nucleoid occlusion is not absolute or (partially) segregated nucleoids are sometimes poorly resolved by fluorescence microscopy.
EnvC is a periplasmic septal ring component
The phenotypes associated with envC mutations suggested that EnvC might be a component of the division apparatus. To determine the subcellular localization of EnvC, we set out to construct a functional fusion to GFP.
EnvC contains two recognizable domains: a large N-terminal domain of ≈ 200 residues predicted to form two or three coiled-coil structures and a C-terminal domain of ≈ 100 residues predicted to belong to the M37 family of metallo-endopeptidases (Bateman et al., 2002; Hara et al., 2002; Ichimura et al., 2002) (Fig. 3B). Based on the annotation of GenBank accession AE000439, the extreme N-terminus of EnvC is predicted to be a _trans_-membrane domain. Ichimura et al. (2002) provided evidence that this is true and that EnvC has an N-out C-in topology. We found, however, that cells expressing an EnvC–GFP fusion that should have contained its supposed _trans_-membrane domain were barely fluorescent, and that the fusion was degraded (data not shown).
The second ATG codon in the GenBank annotation of envC is associated with a more recognizable ribosome binding site (Hara et al., 2002). Using this start, the extreme N-terminus of EnvC is predicted to be a cleavable signal sequence for Sec-mediated export to the periplasm. Counter to the results of Ichimura et al. (2002), Hara et al. (2002) presented evidence that EnvC is processed and that a substantial portion of the processed form is found in the periplasm.
GFP that is exported by the general secretory system folds incorrectly, is not fluorescent and is degraded (Feilmeier et al., 2000). However, prefolded (fluorescent) GFP can be exported to the periplasm by the twin-arginine transport (Tat) system when the signal peptide of the Tat substrate TorA is appended to its N-terminus (Santini et al., 2001; Thomas et al., 2001). In addition, we demonstrated recently that the murein amidases AmiA and AmiC are also Tat substrates and that GFP fusions to these proteins can be used to determine their subcellular localization in the periplasm of live cells (Bernhardt and de Boer, 2003). To localize EnvC in the periplasm, therefore, we constructed an ssAmiC–EnvC–GFP fusion (Fig. 3B), in which the predicted signal sequence of EnvC is replaced by that of AmiC. Immunoblot analysis indicated that, when expressed from a prophage (λTB47), the Tat-targeted fusion (TTEnvC–GFP) remained intact in both wild-type and Δ_envC_ cells (Fig. 6A). Moreover, a significant portion of TTEnvC-GFP was released from spheroplasts, confirming its export to the periplasm (Fig. 6B). Importantly, λTB47 corrected the morphological defects of EnvC– cells and the filamentation phenotype of EnvC– Min– cells, demonstrating that the Tat-exported fusion is functional (Fig. 6C3; data not shown). These results strongly support the proposal that native EnvC is a periplasmic species (Hara et al., 2002), rather than a transmembrane one (Ichimura et al., 2002).
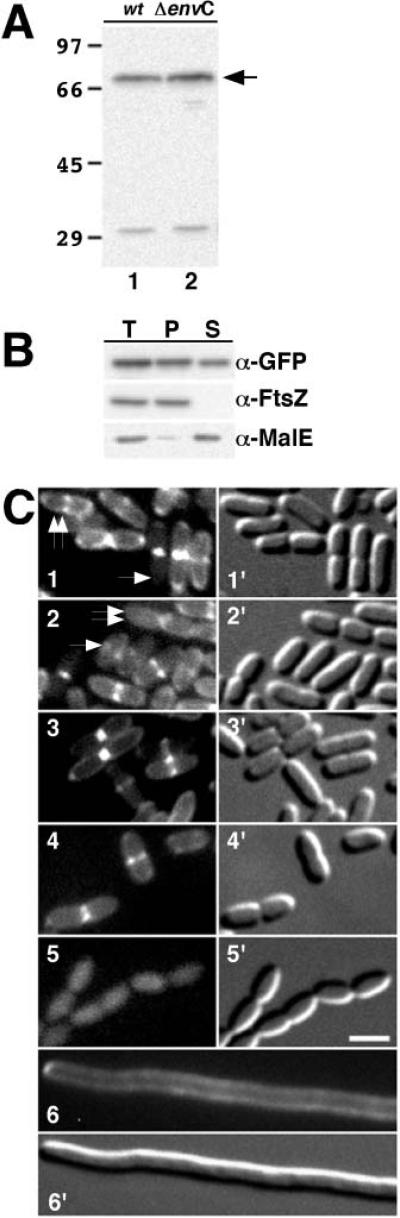
Fig. 6. EnvC is recruited to the septal ring.
A. Immunoblot probed with anti-GFP antibodies. Lanes contained whole-cell extracts of TB28(λTB47) [wt (Plac::ss_amiC-envC-gfp_)] (lane 1) or TB44(λTB47) [Δ_envC_::frt (Plac::ss_amiC-envC-gfp_)] (lane 2). Extracts were prepared from cells grown to an OD600 of 0.5–0.6 in minimal medium supplemented with 50 μM IPTG. The arrow indicates the position of TTEnvC–GFP. Numbers on the left (in kDa) indicate the positions of molecular weight markers.
B. Release of TTEnvC–GFP from spheroplasts. TB28(λTB47) [wt (Plac::ss_amiC-envC-gfp_)] was grown in LB supplemented with 0.2% maltose and 50 μM IPTG. One aliquot of cells was used to prepare a total-cell extract. The remaining cells were converted to spheroplasts and pelleted by centrifugation. The resulting pellet (P) and supernatant (S) fractions, along with the total-cell extract (T), were analysed by SDS–PAGE and immunoblotting for GFP, FtsZ or MalE as indicated. FtsZ and MalE served as markers for the cytoplasm and periplasm respectively.
C. GFP fluorescence (1–6) and DIC (1′-6′) micrographs of live TB28(λTB47) [wt (Plac::ss_amiC-envC-gfp_)] (1 and 2), TB44(λTB47) [Δ_envC_::frt (Plac::ss_amiC-envC-gfp_)] (3), MC4100(λTB47) [wt (Plac::ss_miC-envC-gfp_)] (4), B1LKO(λTB47) [Δ_tatC_ (Plac::ss_amiC-envC-gfp_)] (5) and TB28(λTB47)/pJE80 [wt (Plac::ss_amiC-envC-gfp_)/Para::_sfiA_] (6) cells grown as in (A), except that the culture used for (6) contained 0.2% arabinose and 100 μM IPTG. In (1), the single arrow points to a deeply constricted cell with a bright focus of EnvC–GFP at the septum, and the double arrow points to a cell with a shallow constriction that has a bright EnvC–GFP ring as well as a peripheral signal. In (2), the single arrow points to a small cell with an apparent accumulation of EnvC–GFP signal at mid-cell, and the double arrow points to a very deeply constricted cell with a dominant peripheral signal. This cell has presumably just completed division, and the daughters are about to separate. Bar equals 2 μm.
To sublocalize TTEnvC–GFP, TB28(λTB47) [wt(Plac::ssamiC-envC-gfp)] cells were grown in minimal media supplemented with 50 μM IPTG and analysed by fluorescence microscopy. In the majority of cells, TTEnvC–GFP generated a peripheral fluorescence signal with some bias towards the cell poles. This polar bias is not specific to EnvC as it is typically seen with fused and unfused periplasmic GFP (Santini et al., 2001; Thomas et al., 2001; Bernhardt and de Boer, 2003). Interestingly, about 50% (154/305) of the cells also showed a distinct ring-like accumulation at mid-cell (Fig. 6C1 and 2). Importantly, both peripheral and septal localization patterns of TTEnvC–GFP required a functional Tat system (Fig. 6C4 and 5). In addition, the ring-like accumulation of TTEnvC–GFP was completely eliminated when FtsZ-ring formation was blocked by sfiA expression from pJE80 [Para::sfiA_] (Fig. 6C6). EnvC-ring formation did not appear to require constriction; 79 out of 207 non-constricting cells (38% of class) had a ring-like accumulation of TTEnvC–GFP. The majority of constricting cells (75/98) also had apparent TTEnvC–GFP rings. The ones that did not only showed a peripheral fluorescence signal and were almost all (20/23) cells with very deep constrictions, suggesting that they were just about to separate. Therefore, EnvC does not appear to linger at the nascent poles after the completion of division. Similar results were obtained when TTEnvC–GFP was localized in an Δ_envC mutant under conditions in which the chaining phenotype was corrected by the fusion (Fig. 6C3), showing that the fusion did not require native EnvC for recruitment to the septum.
We conclude that EnvC is recruited to the septal ring, suggesting a direct role for the protein in the division process.
Evidence for an enlarged septal periplasm in EnvC– cells
As mentioned above, expression of ssAmiC–GFP as well as other Tat-targeted GFP constructs (TTGFP) in wild-type cells results in a fairly uniform peripheral fluorescence signal with some bias towards the poles (Santini et al., 2001; Thomas et al., 2001; Bernhardt and de Boer, 2003) (Fig. 7A). Interestingly, however, Δ_envC_ mutants expressing ssAmiC–GFP showed distinct rings of increased fluorescence at mid-cell in approximately half the cells (Fig. 7B). Such rings were never observed in wild-type cells, nor were they specific for ssAmiC fusions; GFP targeted to the periplasm using the Tat signal sequences of TorA or AmiA also produced bands of increased fluorescence at the septa of Δ_envC_ mutants (Fig. 7C and D). As with the presence of constrictions without associated septal rings, the presence of increased amounts of TTGFP at the septa of envC mutants is probably caused by the delay in OM constriction relative to that of the IM (Rodolakis et al., 1973). Such a delay should result in an increased periplasmic volume at septa. When this localized increase is sufficiently large, the concomitant increase in the amount of TTGFP at these sites would appear as rings of increased fluorescence.
Fig. 7.
Evidence for an increased periplasmic volume at the septa of EnvC– cells. GFP fluorescence (A–D) and DIC (A′–D′) micrographs show representative live cells of TB28(λTB46) [wt (Plac::ss_amiC-gfp_)] (A), TB44(λTB46) [Δ_envC_::frt (Plac::ss_amiC-gfp_)] (B), TB44/pTB32 [Δ_envC_::frt/Plac::amiA-gfp)] (C) and TB44(λTB6) [Δ_envC_::frt (Plac::ss_torA-gfp_)] (D) grown in minimal medium supplemented with 500 (A and B), 250 (C) or 50 μM (D) IPTG. Cells were grown to an OD600 of 0.5–0.6. Bar equals 2 μm.
EnvC has murein hydrolytic activity
The C-terminal domain of EnvC belongs to the M37 family of metallo-endopeptidases (Bateman et al., 2002; Hara et al., 2002; Ichimura et al., 2002). Another member of this family is lysostaphin, an enzyme that degrades Staphylococcal murein by cleaving the interpeptide bridges at Gly–Gly bonds (Sloan et al., 1977; Recsei et al., 1987; Hara et al., 2002). Previously, Ichimura et al. (2002) detected a weak endoproteolytic activity of EnvC on a β-casein substrate.
To test whether EnvC also has murein hydrolase activity, we purified a 6×His-tagged derivative of EnvC (EnvC-FKH) and used it in a zymogram assay (Figs 3B and 8). Several dilutions of purified EnvC, along with BSA and egg white lysozyme, were resolved on two identical SDS-polyacrylamide gels impregnated with extensively deproteinized E. coli murein sacculi. One gel was stained with Coomassie blue (Fig. 8A) and the other was incubated overnight in buffer to promote renaturation of the proteins in the gel. The sacculi in the second gel were then stained with methylene blue. In this assay, clear zones indicate destruction of sacculi and, thus, the presence of murein hydrolase activity (Jayaswal et al., 1990). The lysozyme control and all the EnvC dilutions produced significant clear zones in the zymogram while, as expected, the BSA negative control failed to do so (Fig. 8B). The proteases proteinase K and trypsin also failed to produce clear zones in this assay (not shown). The clearing activity of EnvC was reduced significantly by the inclusion of 5 mM EDTA in the renaturation buffer (not shown), suggesting that, as for its weak β-caseinase activity (Ichimura et al., 2002), EnvC may require divalent metal for its murein hydrolase activity.
Fig. 8.
EnvC has murein hydrolytic activity. Coomassie-stained gel (A) and methylene blue-stained zymogram (B) contained molecular weight markers (in descending order: 66, 45, 36, 29, 24, 20 and 14 kDa) (lane 1), 5 μg of BSA (lane 2), 5 μg of egg white lysozyme (Lys) (lane 3) and 5.0, 2.5 or 1.3 μg of purified EnvC–FKH (lanes 4–6). Lys and EnvC–FKH yielded significant clear zones in the zymogram. Minor species running below full-length EnvC–FKH also promoted some clearing. These were probably EnvC–FKH fragments that retained some activity.
We conclude that EnvC possesses murein hydrolase activity and propose that this activity contributes to septal murein cleavage.
Discussion
To identify mutations causing a synthetically lethal or sick phenotype in combination with a defective Min system (slm mutations), we developed a simple, and generally applicable, synthetic lethal screen for E. coli. It was anticipated that a successful screen for slm mutations would yield (novel) genes involved in cell division and shape determination (Yu and Margolin, 2000; Corbin et al., 2002). Accordingly, we isolated a transposon insertion in ybeB, which appears to be polar on the expression of the downstream shape genes pbpA and rodA, and an insertion in envC (yibP), a classical (Rodolakis et al., 1973) but ill-understood division gene. In addition, we recently repeated the screen using a larger mutant library and isolated several more slm mutants. These mutants have interesting division defects and are currently being analysed (T. G. Bernhardt, unpublished results). Our results indicate that, as in yeast, the synthetic lethal methodology can be a powerful tool for uncovering genetic interactions in E. coli.
What causes the Slm phenotypes?
In addition to suffering aberrant polar divisions, Min– cells are mildly filamentous. The classic explanation for this phenotype is that cells only produce enough ‘division potential’ for the formation of one septum per pair of sister chromosomes (Teather et al., 1974; Donachie and Begg, 1996). Thus, non-productive polar divisions occur at the expense of productive mid-cell divisions, leading to a population of elongated cells in addition to normal cells and minicells. Division potential appears to be limited by the levels of FtsA and FtsZ because, when they are both overproduced, Min– cells divide more often and approach a normal length (Begg et al., 1998).
The observation of multiple FtsZ structures in Min– cells indicates that division potential is not all used at one site but is actually diluted over multiple potential division sites (Fig. 4) (Yu and Margolin, 1999). However, the marked differences in fluorescence intensities between the ring structures within the same cell suggest that division potential is not evenly distributed, and that rings are at various stages of maturation. The presence of multiple developing rings in the same cell probably leads to competition for limiting division protein subunits and a consequent delay in the formation of a mature, constriction-competent septal ring. In addition to the frequent misplacement of a septum at a cell pole, such a competition-induced general delay in septal ring maturation is likely to contribute to the mild filamentation phenotype of Min– cells.
Both slm mutations caused a much more severe division defect in Min– cells than in wild-type cells, and the defects were suppressible by overexpression of ftsQAZ in both cases. Therefore, it appears that both slm mutations further reduce the already compromised ability of Min– cells to assemble division-competent septal rings. In the case of _slm_3, this may be directly related to the change in cell shape. It has been proposed that spherical cells require more division proteins to assemble a functional septal ring large enough to accommodate their increased circumference (Vinella et al., 1993). For example, pbpA mutants can only be isolated if the ftsQAZ operon is overexpressed (Vinella et al., 1993; 2000; Navarro et al., 1998). As they are significantly wider than normal, we similarly envisage that _slm_3 cells can ill afford the dilution of division potential caused by a defective Min system. Without Min, the cells grow poorly because they struggle to divide, become elongated and lyse more frequently than normal.
In the case of _slm_11, the cause of the severe filamentation phenotype of EnvC– Min– cells may be subtler. Our results are compatible with a model in which the combined loss of EnvC and the Min system causes a catastrophic reduction in the efficiency of septal ring maturation. In this model, EnvC normally plays a modest role in promoting septal ring maturation, either by stimulating assembly or by counteracting premature disassembly. In Min– cells, where septal ring maturation is already stressed by the dispersion of ring components among multiple structures, EnvC's role in maturation becomes critical. Its loss causes a severe delay in the formation of mature division-competent septal rings, leading to the observed filamentation phenotype and growth defect. In addition to its localization (Fig. 6), a role for EnvC in septal ring maturation is supported by the apparent delay in ring assembly in envC single mutants at 30°C (Fig. 5). Moreover, Ichimura et al. (2002) observed that, at 42°C, the filaments of their insertional yibP mutant contained very few Z-rings, indicating that EnvC's role becomes even more critical at elevated temperatures.
EnvC and septal murein cleavage
Escherichia coli encodes a wide array of periplasmic and OM-bound murein hydrolases (autolysins) with potential access to the murein sacculus: (i) the lytic transglycosylases (sltY, mltA, mltB, mltC, mltD and mltE) (ii) the d,d-endopeptidases (dacB, pbpG and mepA); and (iii) the N-acetylmuramoyl-l-alanine amidases (amiA, amiB and amiC) (Höltje, 1998; Heidrich et al., 2002). The lytic transglycosylases hydrolyse the polysaccharide linkages of murein, while the endopeptidases and amidases can attack the peptide cross-links. A recent analysis of multiple deletion mutants suggests that all these enzymes can participate in splitting the septal murein, but that the amidases play the dominant role in this process (Heidrich et al., 2001; 2002).
Similar to amidase mutants, envC mutants are defective in daughter cell separation, form chains of cells and are sensitive to detergents (Rodolakis et al., 1973; Hara et al., 2002; Heidrich et al., 2002; Ize et al., 2003) (Figs 2B5 and 4B). Electron microscopy studies indicate that the phenotypes of both mutants are associated with a defect in septal murein splitting, causing a delay in the constriction of the OM (Rodolakis et al., 1973; Heidrich et al., 2001; 2002). Compared with Ami– chains, the constrictions we observed in EnvC– chains tended to be shallower, suggesting that the delay in OM invagination may be more severe in the latter. This delay probably results in the presence of constriction sites without associated septal rings, and in what appears to be an enlarged periplasm at the septa of envC mutants (Figs 4B, 5 and 7).
Our results, together with those of Hara et al. (2002), indicate that EnvC participates directly in the splitting process. It is in the right compartment (the periplasm), at the right place (the septum) and has the necessary activity (the ability to hydrolyse murein) (Figs 6 and 8). It will be interesting to establish what specific murein linkages are cleaved by EnvC. Its weak endoproteolytic activity on β-casein (Ichimura et al., 2002) and its similarity to the endopeptidase lysostaphin (Bateman et al., 2002; Hara et al., 2002) suggest that it attacks the peptide cross-links. Further work will be required to determine whether this is indeed the case. How EnvC is recruited to the division apparatus, and how its murein hydrolytic activity might be related to its proposed role in the assembly and/or stability of the septal ring are other interesting questions that deserve further investigation. The presence of a coiled-coil region in EnvC is intriguing. Two of the _trans_-membrane septal ring proteins, FtsB and FtsL, contain predicted coiled-coils in their periplasmic domains (Buddelmeijer et al., 2002), and it is conceivable that EnvC interacts with one or both of these.
Proteins containing M37 domains are found throughout the bacterial domain (Bateman et al., 2002). Besides EnvC, E. coli encodes three other proteins with a lysostaphin-like M37 domain: NlpD, YebA and YgeR (SWISSPROT P33648, P24204 and Q46798 respectively). Of these, only the lipoprotein NlpD has been characterized to some extent. No morphological phenotypes have been reported for nlpD mutants (Li and Clarke, 1992; Ichikawa et al., 1994). However, overexpression of NlpD results in lysis, suggesting that it might also be a murein hydrolase (Lange and Hengge-Aronis, 1994). Additional work on this class of proteins is likely to shed new light on the growth and (re)modelling of the murein sacculus during the E. coli cell cycle.
Artificial Tat targeting of GFP fusions
We recently used functional periplasmic GFP fusions to demonstrate that the amidases AmiA and AmiC are natural substrates of the Tat export pathway. This analysis also revealed that, although AmiA appears to be located throughout the periplasm at all stages of growth, AmiC is recruited to the septal ring at the onset of constriction (Bernhardt and de Boer, 2003). Here, we extended this approach by routing a GFP fusion to what is most probably a natural Sec substrate, through the Tat system, to study its periplasmic localization. This allowed us to classify EnvC as another periplasmic autolysin that is recruited to the septal ring. The use of such TTGFP fusions is likely to be a viable approach for sublocalizing many other exported proteins.
Experimental procedures
Media, bacterial strains, plasmids and phages
Cells were grown in LB (1% tryptone, 0.5% yeast extract and 0.5% NaCl) or minimal M9 media (Miller, 1972) supplemented with 0.2% maltose, 0.2% casamino acids and 50 μM thiamine. Where appropriate, antibiotics were used at 12.5 (Tet), 25 (Kan, Cam) or 50 (Amp) μg μl−1.
Bacterial strains used in this study are listed in Table 1. The alleles lacIZYA < >aph, minCDE < >aph, envC < >aph and PminC < >aph araC Para were constructed by λRed-mediated recombineering (Datsenko and Wanner, 2000; Yu et al., 2000) and then moved into the desired strains by P1-mediated transduction. The symbol < > denotes DNA replacement (Yu et al., 2000), and frt denotes a scar sequence remaining after eviction of the aph (KanR) cassette by FLP recombinase (Datsenko and Wanner, 2000).
Table 1.
Bacterial strains used in this study.
| Strain | Relevant genotypea | Source or reference |
|---|---|---|
| BL21(λDE3) | ompT rB– mB– (PlacUV5::T7_gene1_) | Novagen |
| MG1655 | _rph_1 _ilvG rfb_-50 | Guyer et al. (1981) |
| DY329 | rph_1 IN(rrnD-rrnE) Δ(argF-lac)U169_nadA::Tn_10_ _gal_490 λ_cI_857 Δ(cro-bioA) | Yu et al. (2000) |
| MC4100 | _araD_139 Δ(argF-lac)U169 _rpsL_150 _relA_1 _flbB_5301 _deoC_1 _ptsF_25 rbsR | Casadaban (1976) |
| B1LKO | MC4100 Δ_tatC_ | Bogsch et al. (1998) |
| DX1 | _dadR_1 _trpE_61 trpA_62 tna_-5 Δ_minB::kan recA::Tn_10 | de Boer et al. (1991) |
| DH5α | F– _hsdR_17 _deoR recA_1 _endA_1 _phoA supE_44 _thi_-1 _gyrA_96 _relA_1 Δ(lacZYA-argF)U169 ϕ80d_lacZ_ΔM15 | Gibco BRL |
| TB12 | MG1655 laclZYA < >aph | Bernhardt and de Boer (2003) |
| TB13 | MG1655 laclZYA < >frt | This study |
| TB14 | TB13 minCDE < >aph | This study |
| TB15 | TB13 minCDE < >frt | This study |
| Slm3 | TB15 _slm_3 (ybeB::EZTnKan-2) | This study |
| Slm11 | TB15 _slm_11(envC::EZTnKan-2) | This study |
| TB27 | TB13 envC < >aph | This study |
| TB28 | MG1655 laclZYA < >frt | Bernhardt and de Boer (2003) |
| TB35 | TB28 envC < >aph | This study |
| TB44 | TB28 envC < >frt | This study |
| TB53 | TB28 amiA::cat amiC::aph | Bernhardt and de Boer (2003) |
| TB55 | DY329 PminC < >(aph araC Para) | This study |
| TB56 | TB28 PminC < >(aph araC Para) | This study |
| TB57 | TB28 PminC < >(frt araC Para) | This study |
| TB58 | TB57 envC < >aph | This study |
| TB61 | TB57 recA::Tn_10_ | This study |
| TB62 | TB58 recA::Tn_10_ | This study |
| TB75 | TB28 _slm_3 (ybeB::EZTnKan-2) | This study |
| TB81 | TB57 _slm_3 (ybeB::EZTnKan-2) | This study |
Salient features of plasmids and phages are indicated in Fig. 3 and the text. In all cases where gfp was used, the allele is _gfpmut_2 (Cormack et al., 1996). Plasmids pJE80 [cat araC Para::_sfiA_] (Johnson et al., 2002), pCH151 [bla lacIq Plac::_zipA-gfp_] and pTB32 [bla lacIq Plac::_amiA-gfp_] (Bernhardt and de Boer, 2003) have been described before. Except for the mini-F derivative pTB8 [bla lacIq Plac::_minCDE lacZ_], pTB57 (see below) and the pSC101 derivative pTB63 [_tet ftsQAZ_], all the plasmids constructed for this study are derivatives of the medium-copy vector pMLB1113 [bla lacIq Plac::_lacZ_] (de Boer et al., 1989) and carry the indicated gene(s) downstream from the lac promoter.
Phages λCH151 [bla lacIq Plac::_zipA-gfp_], λTB6 [bla lacIq Plac::ss_torA-gfp_], λTB46 [bla lacIq Plac::ss_amiC-gfp_] and λTB47 [bla lacIq Plac::ss_amiC-envC-gfp_] were obtained by crossing lNT5 [_imm_21] with the appropriate pMLB1113 derivative (de Boer et al., 1989). λTB47 encodes a Tat-targeted version of EnvC–GFP in which the first 34 residues of EnvC were replaced by the signal sequence of AmiC (residues 1–31) (Bernhardt and de Boer, 2003).
Further details on the construction of strains, plasmids and phages are provided in Supplementary material.
Transposon mutagenesis, screening for slm mutants and mapping insertion sites
Electrocompetent cells of TB15/pTB8 [Δ_lacIZYA_ Δ_minCDE_/bla Plac::_minCDE lacZ_] were prepared using ice-cold 10% glycerol as described previously (Dower et al., 1988). Aliquots (40 μl) were mixed with 1 μl (20 ng) of the EZTnKan-2 transposome from Epicentre. After electroporation, the cells were outgrown for 2 h at 30°C in LB-Amp supplemented with 500 μM IPTG, plated on LB-Amp-Kan agar supplemented with 500 μM IPTG and grown overnight at 30°C. Five separate electroporations yielded a total of about 10 000 colonies. These colonies were resuspended in 5 ml of LB, to which 3 ml of 40% glycerol was added. This library of mutants was aliquoted and stored at –80°C.
For the screen, an aliquot of the library was thawed, a 5 × 10−7 dilution was prepared in LB, and 0.1 ml aliquots were spread on LB agar plates supplemented with 500 μM IPTG and 60 μg ml−1 Xgal (LB-IX agar). To avoid selection for cells in which the transposon had inserted in the plasmid, kanamycin was omitted from the medium at this step. The plates were incubated at 30°C for 2 days so that most of the colonies were large enough to see the sectored-blue appearance of the Xgal staining clearly (Fig. 1A). Each plate contained ≈100 colonies, and we screened about 30 000 colonies for the atypical ones that appeared to be solid-blue. These were purified on LB-Amp-Kan-IPTG (500 μM) and tested for growth and colony phenotypes on LB-IX and LB–glucose agar.
The positions of transposon insertions were determined using arbitrary PCR essentially as described previously (O'Toole and Kolter, 1998). More details on this procedure are provided in Supplementary material.
Purification of EnvC and zymography
Plasmid pTB57 [PT7::_envC-fkh_] (Fig. 3B) is a pET21b (Novagen) derivative and encodes a non-exported derivative of EnvC in which its native signal sequence (residues 1–34) is replaced by the tripeptide MAS, and its C-terminus is fused to the FKH-tag peptide MDYKDDDDKARRASVEFHIE(H)6 (Hale et al., 2000). EnvC-FKH was overproduced in BL21(lDE3)/pTB57 cells and purified using Ni2+-affinity chromatography essentially as described previously (Lackner et al., 2003). Details of the purification procedure can be found in Supplementary material.
Zymography was performed essentially as described previously (Abanes-De Mello et al., 2002) using extensively deproteinized TB28 sacculi prepared as described previously (de Pedro et al., 1997). Two SDS–PAGE gels (14%T, 2.7%C), containing 0.6% (wet w/v) sacculi, were run concurrently with lanes loaded as indicated in Fig. 8. One gel was fixed and stained with Coomassie Brilliant Blue. The other was incubated overnight at room temperature in renaturation buffer (25 mM Tris-HCl, 1% TritonX-100, pH 8.0), stained with 0.1% methylene blue in 0.01% KOH for 3 h and destained with H2O.
Microscopy and other methods
Cultures were grown at 30°C. For the micrographs shown in Figs 2B and 4, cells from a saturated culture grown in LBAmp-IPTG (500 μM) or LB-arabinose (0.2%), respectively, were diluted 1:400 into LB-Amp-IPTG (500 μM) (Fig. 2B1–3 and 2B5), LB-Amp (Fig. 2B2, 2B4 and 2B6) or LB-IPTG (50 μM) (Fig. 4) and grown to an OD600 between 0.6 and 0.8. The cells were then fixed and visualized as described below. See the figure legends for the specific concentrations of IPTG and arabinose used to supplement other cultures. For the micrographs in Figs 6C and 7, cells from a saturated culture were diluted 1:100 in M9 medium, grown to an OD600 between 0.5 and 0.6 and visualized live. For the spheroplasting experiment in Fig. 6B, cells from a saturated culture were diluted 1:100 into LB and grown to an OD600 of ≈ 0.6.
GFP fluorescence and DIC microscopy were performed essentially as described previously (Johnson et al., 2002). Where appropriate, cells in LB were fixed as described previously (Johnson et al., 2002), except that the fixation was carried out at room temperature for 30 min.. For DAPI staining of nucleoids, fixed cells were treated with 0.25 μg ml−1 DAPI and viewed immediately using a DAPI filter set (395 nm dichroic mirror, 359–371 nm excitation filter and a 385 nm longpass barrier filter).
Cell lengths and the positions of constrictions and ZipA–Gfp rings were measured using OBJECT IMAGE (Vischer et al., 1994). Immunoblotting, spheroplasting and the preparation of whole-cell extracts were performed exactly as described previously (Bernhardt and de Boer, 2003).
Supplementary Material
sm
Acknowledgements
We thank Felipe Bendezu, Don Court, Cynthia Hale, Jay Johnson, Tracy Palmer, David Raskin, Phil Rather, Barry Wanner and Seiichi Yasuda for strains and plasmids, and members of our laboratory for support and helpful comments. This work was supported by NIH grant GM57059. Thomas G. Bernhardt is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1698-02).
Footnotes
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerlund T, Bernander R, Nordström K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, et al. The Pfam protein families data-base. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K, Nikolaichik Y, Crossland N, Donachie WD. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Hand AR, Rothfield LI. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Judson N, Boyd D, Mekalanos JJ, Beckwith J. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc Natl Acad Sci USA. 2002;99:6316–6321. doi: 10.1073/pnas.092128499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Corbin BD, Yu XC, Margolin W. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 2002;21:1998–2008. doi: 10.1093/emboj/21.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie WD, Begg KJ. Division potential in Escherichia coli. J Bacteriol. 1996;178:5971–5976. doi: 10.1128/jb.178.20.5971-5976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hale CA, de Boer PAJ. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ, and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PAJ. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J Bacteriol. 2002;184:2552–2556. doi: 10.1128/JB.184.9.2552-2556.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Rhee AC, de Boer PAJ. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Meinhardt H, de Boer PAJ. Dynamic localization cycle of the cell division regulator MinE in E. coli. EMBO J. 2001;20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CD, Coe EH, Jr, Martienssen RA. Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO J. 1992;11:4037–4046. doi: 10.1002/j.1460-2075.1992.tb05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Narita S, Karibian D, Park JT, Yamamoto Y, Nishimura Y. Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiol Lett. 2002;212:229–236. doi: 10.1111/j.1574-6968.2002.tb11271.x. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, Höltje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Rutenberg AD, de Vet S. Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys Rev Lett. 2001;87:278102. doi: 10.1103/PhysRevLett.87.278102. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci USA. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Saez C, Lutkenhaus J. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J Bacteriol. 2003;185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Meir Y, Wingreen NS. Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc Natl Acad Sci USA. 2003;100:12724–12728. doi: 10.1073/pnas.2135445100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa JK, Li C, Fu J, Clarke S. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Yamazoe M, Maeda M, Wada C, Hiraga S. Proteolytic activity of YibP protein in Escherichia coli. J Bacteriol. 2002;184:2595–2602. doi: 10.1128/JB.184.10.2595-2602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- Jayaswal RK, Lee YI, Wilkinson BJ. Cloning and expression of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. J Bacteriol. 1990;172:5783–5788. doi: 10.1128/jb.172.10.5783-5788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Lackner LL, de Boer PAJ. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ-rings. J Bacteriol. 2002;184:2951–2962. doi: 10.1128/JB.184.11.2951-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Lackner LL, Hale CA, de Boer PAJ. ZipA is required for targeting of DMinC/DicB-, but not DMinC/MinD-, complexes to septal ring assemblies in Escherichia coli. J Bacteriol. 2004 doi: 10.1128/JB.186.8.2418-2429.2004. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop AH, Hartley ME, Bourgeois S. A low-copy-number vector utilizing β-galactosidase for the analysis of gene control elements. Gene. 1987;52:245–256. doi: 10.1016/0378-1119(87)90051-5. [DOI] [PubMed] [Google Scholar]
- Kruse K. A dynamic model for determining the middle of Escherichia coli. Biophys J. 2002;82:618–627. doi: 10.1016/S0006-3495(02)75426-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Raskin DM, de Boer PA. ATP–dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol. 2003;185:735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol Microbiol. 1994;13:733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- Li C, Clarke S. A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat-shock resistance. Proc Natl Acad Sci USA. 1992;89:9885–9889. doi: 10.1073/pnas.89.20.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, de Boer PAJ. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci USA. 2001;98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Mulder E, Woldringh CL. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Robin A, D'Ari R, Joseleau-Petit D. Analysis of the effect of ppGpp on the ftsQAZ operon in Escherichia coli. Mol Microbiol. 1998;29:815–823. doi: 10.1046/j.1365-2958.1998.00974.x. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje J-V, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J Bacteriol. 2001;183:6630–6635. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PAJ. MinDE dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999a;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PAJ. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999b;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei PA, Gruss AD, Novick RP. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci USA. 1987;84:1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolakis A, Thomas P, Starka J. Morphological mutants of Escherichia coli. Isolation and ultrastructure of a chain-forming envC mutant. J Gen Microbiol. 1973;75:409–416. doi: 10.1099/00221287-75-2-409. [DOI] [PubMed] [Google Scholar]
- Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J Biol Chem. 2001;276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan GL, Smith EC, Lancaster JH. Lysostaphin endopeptidase-catalysed transpeptidation reactions of the imino-transfer type. Biochem J. 1977;167:293–296. doi: 10.1042/bj1670293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG, Boyd A, Stoker N. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J Bacteriol. 1980;143:569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suefuji K, Valluzzi R, RayChaudhuri D. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc Natl Acad Sci USA. 2002;99:16776–16781. doi: 10.1073/pnas.262671699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Yu X-C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- Teather RM, Collins JF, Donachie WD. Quantal behaviour of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- Vinella D, Joseleau-Petit D, Thévenet D, Bouloc P, D'Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J Bacteriol. 1993;175:6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Cashel M, D'Ari R. Selected amplification of the cell division genes ftsQ-ftsA-ftsZ. Escherichia coli. Genetics. 2000;156:1483–1492. doi: 10.1093/genetics/156.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vischer NOE, Huls PG, Woldringh CL. Object-Image: an interactive image analysis program using structured point collection. Binary. 1994;6:160–166. [Google Scholar]
- Yu X-C, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- Yu X-C, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J Bacteriol. 2000;182:6203–6213. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sm