Orchestrating high-throughput genomic analysis with Bioconductor (original) (raw)
. Author manuscript; available in PMC: 2015 Jul 21.
Published in final edited form as: Nat Methods. 2015 Feb;12(2):115–121. doi: 10.1038/nmeth.3252
Abstract
Bioconductor is an open-source, open-development software project for the analysis and comprehension of high-throughput data in genomics and molecular biology. The project aims to enable interdisciplinary research, collaboration and rapid development of scientific software. Based on the statistical programming language R, Bioconductor comprises 934 interoperable packages contributed by a large, diverse community of scientists. Packages cover a range of bioinformatic and statistical applications. They undergo formal initial review and continuous automated testing. We present an overview for prospective users and contributors.
1 Introduction
Progress in biotechnology is continually leading to new types of data, and the data sets are rapidly increasing in volume, resolution and diversity. This promises unprecedented advances in our understanding of biological systems and in medicine. However, the complexity and volume of data also challenge scientists’ ability to analyze them. Meeting this challenge requires continuous improvements in analysis tools and associated software engineering.
Bioconductor [1] provides core data structures and methods that enable genome-scale analysis of high-throughput data in the context of the rich statistical programming environment offered by the R project [2]. It supports many types of high-throughput sequencing data (including DNA, RNA, chromatin immunoprecipitation, Hi-C, methylomes and ribosome profiling) and associated annotation resources; contains mature facilities for microarray analysis [3]; and covers proteomic, metabolomic, flow cytometry, quantitative imaging, cheminformatic and other high-throughput data. Bioconductor enables the rapid creation of workflows combining multiple data types and tools for statistical inference, regression, network analysis, machine learning and visualization at all stages of a project from data generation to publication.
Bioconductor is also a flexible software engineering environment in which to develop the tools needed, and it offers users a framework for efficient learning and productive work. The foundations of Bioconductor and its rapid coevolution with experimental technologies are based on two motivating principles.
The first is to provide a compelling user experience. Bioconductor documentation comes at three levels: workflows that document complete analyses spanning multiple tools; package vignettes that provide a narrative of the intended uses of a particular package, including detailed executable code examples; and function manual pages with precise descriptions of all inputs and outputs together with working examples. In many cases, users ultimately become developers, making their own algorithms and approaches available to others.
The second is to enable and support an active and open scientific community developing and distributing algorithms and software in bioinformatics and computational biology. The support includes guidance and training on software development and documentation, as well as the use of appropriate programming paradigms such as unit testing and judicious optimization. A primary goal is the distributed development of interoperable software components by scientific domain experts. In part we achieve this by urging the use of common data structures that enable workflows integrating multiple data types and disciplines. To facilitate research and innovation, we employ a high-level programming language. This choice yields rapid prototyping, creativity, flexibility and reproducibility in a way that neither point-and-click software nor a general-purpose programming language can. We have embraced R for its scientific and statistical computing capabilities, for its graphics facilities and for the convenience of an interpreted language. R also interfaces with low-level languages including C and C++ for computationally intensive operations, Java for integration with enterprise software and JavaScript for interactive web-based applications and reports.
2 The user perspective
The Bioconductor user community is large and international (Table 1). Users benefit from the project in different ways. A typical encounter with Bioconductor (Box 1) starts with a specific scientific need, for example, differential analysis of gene expression from an RNA-seq experiment. The user identifies the appropriate documented workflow, and because the workflow contains functioning code, the user runs a simple command to install the required packages and replicate the analysis locally. From there, she proceeds to adapt the workflow to her particular problem. To this end, additional documentation is available in the form of package vignettes and manual pages. She can load further packages with additional functionality. When help is needed, the user can turn to the support forum with questions on the software and the underlying science, and she can attend training courses and conferences. Some users move from using to developing software, and Bioconductor encourages this transition.
Table 1.
Usage and impact-related statistics.
| Metric | Statistic |
|---|---|
| Unique IP addresses downloading Bioconductor packages | 323,119 within the last 12 months |
| Support site and/or mailing list contributors | 1,331 within the last 12 months |
| Support site visitors | >8,200 unique per month |
| Developer mailing list subscribers | 927 |
| Full-text articles at PubMed Central mentioning Bioconductor | 10,642 |
| PubMed-indexed articles citing Bioconductor packages | 22,838 |
| TCGA Consortium papers using Bioconductor tools | at least 12 of 15 |
2.1 Case study: high-throughput sequencing data analysis
Analysis of large-scale RNA or DNA sequencing data often begins with aligning reads to a reference genome, which is followed by interpretation of the alignment patterns. Alignment is handled by a variety of tools, whose output typically is delivered as a BAM file. The Bioconductor packages Rsamtools and GenomicAlignments provide a flexible interface for importing and manipulating the data in a BAM file, for instance for quality assessment, visualization, event detection and summarization.
The regions of interest in such analyses are genes, transcripts, enhancers or many other types of sequence intervals that can be identified by their genomic coordinates. Bioconductor supports representation and analysis of genomic intervals with a “Ranges” infrastructure that encompasses data structures, algorithms and utilities including arithmetic functions, set operations and summarization [4] (Fig. 1). It consists of several packages including IRanges, GenomicRanges, GenomicAlignments, GenomicFeatures, VariantAnnotation and rtracklayer. The packages are frequently updated for functionality, performance and usability. The Ranges infrastructure was designed to provide tools that are convenient for end users analyzing data while retaining flexibility to serve as a foundation for the development of more complex and specialized software. We have formalized the data structures to the point that they enable interoperability, but we have also made them adaptable to specific use cases by allowing additional, less formalized user-defined data components such as application-defined annotation.
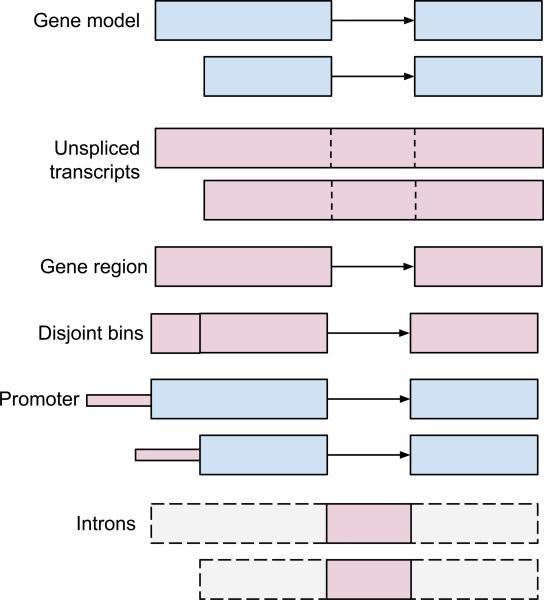
Figure 1.
Example uses of the Ranges algebra. A GRanges object, g (top), represents two transcript isoforms of a gene, each with two exons. The coordinates of unspliced transcripts are identified with the function range(g). Calculating the gene region involves flattening the gene model into its constituent exons and reducing these to nonoverlapping ranges, reduce(unlist(g)). Ranges defining disjoint bins, disjoin(unlist(g)), are useful in counting operations, e.g., in RNA-seq analysis. Putative promoter ranges are found using strand-aware range extension, flank(range(g), width=100). Elementary operations can be composed to succinctly execute queries such as psetdiff(range(g), g) for computing the intron ranges.
Workflows can differ vastly depending on the specific goals of the investigation, but a common pattern is reduction of the data to a defined set of ranges in terms of quantitative and qualitative summaries of the alignments at each of the sites. Examples include detecting coverage peaks or concentrations in chromatin immunoprecipitation–sequencing, counting the number of cDNA fragments that match each transcript or exon (RNA-seq) and calling DNA sequence variants (DNA-seq). Such summaries can be stored in an instance of the class GenomicRanges.
2.2 Coordinated analysis of multiple samples
To facilitate the analysis of experiments and studies with multiple samples, Bioconductor defines the SummarizedExperiment class. The computed summaries for the ranges are compiled into a rectangular array whose rows correspond to the ranges and whose columns correspond to the different samples (Fig. 2). For a typical experiment, there can be tens of thousands to millions of ranges and from a handful to hundreds of samples. The array elements do not need to be single numbers: the summaries can be multivariate.
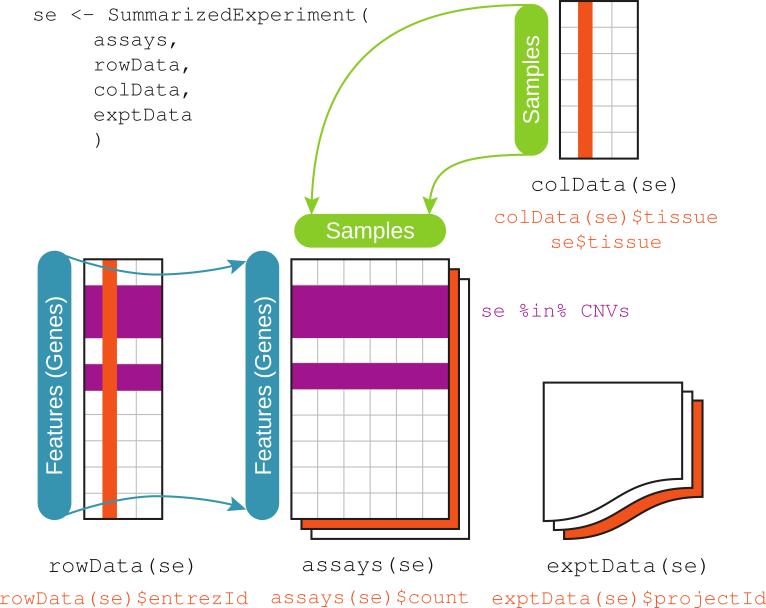
Figure 2.
The integrative data container SummarizedExperiment. Its assays component is one or several rectangular arrays of equivalent row and column dimensions. Rows correspond to features, and columns to samples. The component rowData stores metadata about the features, including their genomic ranges. The colData component keeps track of sample-level covariate data. The exptData component carries experiment-level information, including MIAME (minimum information about a microarray experiment)-structured metadata [21]. The R expressions exemplify how to access components. For instance, provided that these metadata were recorded, rowData(se)$entrezId returns the NCBI Entrez Gene identifiers of the features, and se$tissue returns the tissue descriptions for the samples. Range-based operations, such as %in%, act on the rowData to return a logical vector that selects the features lying within the regions specified by the data object CNVs. Together with the bracket operator, such expressions can be used to subset a SummarizedExperiment to a focused set of genes and tissues for downstream analysis.
The SummarizedExperiment class also stores metadata on the rows and columns. Metadata on the samples usually include experimental or observational covariates as well as technical information such as processing dates or batches, file paths, etc. Row metadata comprise the start and end coordinates of each feature and the identifier of the containing polymer, for example, the chromosome name. Further information can be inserted, such as gene or exon identifiers, references to external databases, reagents, functional classifications of the region (e.g., from efforts such as the Encyclopedia of DNA Elements (ENCODE) [5]) or genetic associations (e.g., from genome-wide association studies, the study of rare diseases, or cancer genetics). The row metadata aid integrative analysis, for example, when matching two experiments according to overlap of genomic regions of interest. Tight coupling of metadata with the data reduces opportunities for clerical errors during reordering or subsetting operations.
2.3 Annotation packages and resources
Reference genomes, annotations of genomic regions and associated gene products (transcripts or proteins), and mappings between molecule identifiers are essential for placing statistical and bioinformatic results into biological perspective. These needs are partly addressed by the Bioconductor annotation data repository, which provides 894 prebuilt standardized annotation packages for use with common model organisms as well as other organisms. Each of the packages presents its data through a standard interface using defined Bioconductor classes, including classes for whole-genome sequences (BSgenome), gene model or transcript databases (TxDb) derived from UCSC (University of California, Santa Cruz) tracks or BioMart annotations, and identifier cross-references from the US National Center for Biotechnology Information, or NCBI (org). There are also facilities for users to create their own annotation packages.
The AnnotationHub resource provides ready access to more than 10,000 genome-scale assay and annotation data sets obtained from Ensembl, ENCODE, dbSNP, UCSC and other sources and delivered in an easy-to-access format (e.g., Ranges-compatible, where appropriate). Bioconductor also supports direct access to underlying file formats such as GTF, 2bit or indexed FASTA.
Bioconductor also offers facilities for directly accessing online resources through their application programming interfaces. This can be valuable when a resource is not represented in an annotation package or when the very latest version of the data is required. The rtracklayer package accesses tables and tracks underlying the UCSC Genome Browser, and the biomaRt package supports fine-grained on-line harvesting of Ensembl, UniProt, COSMIC (Catalogue Of Somatic Mutations In Cancer) and allied resources. Many additional packages access web resources, for example, KEGGREST, PSICQUIC and Uniprot.ws.
2.4 Experiment data packaging and access
The Bioconductor experiment data repository currently contains 224 packages. Experiment data packages have important roles as example data sets on which methods are demonstrated; some can be used for benchmarking and comparing methods, and some are reproducible descriptions of analyses reported in scientific papers (e.g., the data and vignette of the package Hiiragi2013 reproduce a recently reported transcriptome analysis of single cells in mouse embryos [6]). Archives of published experiment data can be harvested using the GEOquery, ArrayExpress and SRAdb packages.
2.5 Integrative analysis
High-throughput assays such as sequencing, flow cytometry [7] and mass spectrometry continue to decrease in cost and increase in quality. Analyses comprising several assays on the same set of samples are becoming more common. Integrative analysis of multiple data types is perhaps the least standardizable task in genomic data analysis, where the need for a flexible working environment in a high-level language such as R is most apparent.
Integrative analysis of multiple assays generally relies on linking through genomic location or annotation. This includes associating genomic locations with transcripts and protein sequences, proteins with other gene products that function in the same pathway or process, and many other possible associations. The combined computation on multiple linked data types and annotations is the essence of integrative genomic analysis.
To perform such analyses, one must use compatible systems of identifiers, reference genomes, gene models, coordinate systems, and so on. For instance, the identification of RNA-editing sites requires that the user have an accurate genotype for the individual as well as RNA-seq reads aligned to that genotype, and variant calls from a DNA-seq experiment should retain not only information about the alignment software but also the precise version of the genome that was used. Bioconductor software is intended to make it easy and automatic to keep track of such issues. This also helps other analysts to determine whether and how a particular processed data set can be integrated with other data sets.
2.6 Visualization
Visualization is essential to genomic data analysis. We distinguish among three main scenarios, each having different requirements. The first is rapid interactive data exploration in “discovery mode”. The second is the recording, reporting and discussion of initial results among research collaborators, often done via web pages with interlinked plots and tool-tips providing interactive functionality. Scripts are often provided alongside to document what was done. The third is graphics for scientific publications and presentations that show essential messages in intuitive and attractive forms. The R environment offers powerful support for all these flavors of visualization—using either the various R graphics devices or HTML5-based visualization interfaces that offer more interactivity—and Bioconductor fully exploits these facilities. Visualization in practice often requires that users perform computations on the data, for instance, data transformation and filtering, summarization and dimension reduction, or fitting of a statistical model. The needed expressivity is not always easy to achieve in a point-and-click interface but is readily realized in a high-level programming language. Moreover, many visualizations, such as heat maps or principal-component analysis plots, are linked to mathematical and statistical models—for which access to a scientific computing library is needed.
A genomics-specific visualization type is plots along genomic coordinates. There are several packages that create attractive displays of along-genome data tracks, including Gviz and ggbio (Fig. 3). These packages operate directly on common Bioconductor data structures and thus integrate with available data manipulation and modeling functionality. A basic operation underlying such visualizations is computing with genomic regions, and the biovizBase package provides a bridge between the Ranges infrastructure and plotting packages. Direct communication between R and genome browsers is implemented by the rtracklayer (for the UCSC Genome Browser) and SRAdb (for the Integrative Genomics Viewer) packages. The epivizr package implements interactive visualization of user data within a lightweight genome browser [8].
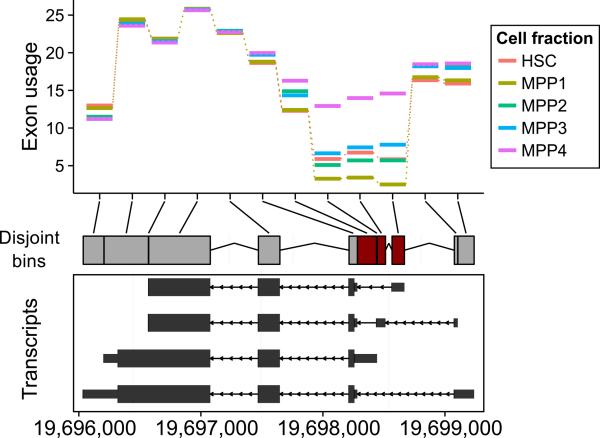
Figure 3.
Visualization along genomic coordinates with ggbio. The plot shows the gene Apoe alongside RNA-seq data from mouse hematopoietic stem cells (HSC) and four fractions of multipotent progenitor (MPP) cells [22]. The disjoint bins (center) were computed from the four transcript isoforms shown in the bottom panel. The y axis of the top panel shows the relative exon usage coefficients estimated with the DEXSeq method [23]. Regions detected as differentially used between the cell fractions are colored dark red in the center panel.
Genomic data set sizes sometimes exceed what can be managed with standard in-memory data models, and then tools from high-performance computing come into play. An example is the use of _rhdf5_—an interface to the HDF5 large data management system (http://www.hdfgroup.org/HDF5)—by the h5vc package to slice large, genome-size data cubes into chunks that are amenable for rapid interactive computation and visualization. Both ggbio and Gviz issue range-restricted queries to file formats including BAM, BGZIP/Tabix and BigWig via Rsamtools and rtracklayer to quickly integrate data from multiple files over a specific genomic region.
2.7 Reproducible research
It can be surprisingly difficult to retrace the computational steps performed in a genomics research project. One of the goals of Bioconductor is to help scientists report their analyses in a way that allows exact recreation by a third party of all computations that transform the input data into the results, including figures, tables and numbers [9]. The project's contributions comprise an emphasis on literate programming vignettes, the BiocStyle and ReportingTools packages, the assembly of experiment data and annotation packages, and the archiving and availability of all previously released packages. A number of developments in the wider R community, including the knitr and rmarkdown packages and the integrated development environment RStudio, make it easy to author attractive vignettes. In addition to the traditional delivery format as a PDF file, the newer generation of tools allow use of HTML5 facilities for interactive visualization, including ‘drill-down’ to expand the view on a specific detail, faceted filtering and comprehensive hyperlinking. Full remote reproducibility remains a challenging problem, in particular for computations that require large computing resources or access data through infrastructure that is potentially transient or has restricted access (e.g., the cloud). Nevertheless, many examples of fully reproducible research reports have been produced with Bioconductor [6,10–14].
2.8 Alternative and complementary tools
Using Bioconductor requires a willingness to modify and eventually compose scripts in a high-level computer language, to make informed choices between different algorithms and software packages, and to learn enough R to do the unavoidable data wrangling and troubleshooting. Alternative and complementary tools exist; in particular, users may be ready to trade some loss of flexibility, automation or functionality for simpler interaction with the software, such as by running single-purpose tools or using a point-and-click interface.
Workflow and data management systems such as Galaxy [15] and Illumina BaseSpace provide a way to assemble and deploy easy-to-use analysis pipelines from components from different languages and frameworks. The IPython notebook [16] provides an attractive interactive workbook environment. Although its origins are with the Python programming language, it now supports many languages, including R. In practice, many users will find a combination of platforms most productive for them.
3 The developer perspective
3.1 The package ecosystem
All software distributed by Bioconductor is in the form of R packages. This simplifies software delivery, use and maintenance, but it puts a burden on the developers. They need to learn how to write R packages, including documentation and test cases (Box 2).
Developers are constantly updating their packages to extend capabilities, improve performance, fix bugs and enhance documentation. These changes are introduced into the development branch of Bioconductor and released to end users every 6 months; changes are tracked using a central, publicly readable Subversion software repository, so details of all changes are fully accessible. Simultaneously, R itself is continually changing, typically around performance enhancements and increased functionality. Owing to this dynamic environment, all packages undergo a daily testing procedure. Testing is fully automated and ensures that all code examples in the package documentation, as well as further unit tests, run without error. Successful completion of the testing will result in the package being built and presented to the community.
Many Bioconductor packages have extensive code examples and tests with which the authors can ensure that their software remains functional even as components up- and downstream change. Of equal importance is keeping the documentation synchronized with changes in the code. Although the testing system places a substantial load both on the central repository and on all developers, it provides a degree of software coherence and usability that is rare in software projects with a diverse and distributed developer community. Of course, limitations exist: the reach and stringency of the tests, beyond the required minimum, vary depending on the package authors. The quality of the repository was highlighted in an editorial in Nature Genetics [17], which listed the Comprehensive R Archive Network (CRAN) and Bioconductor as the only software repositories endorsed by that journal, across all programming languages.
3.2 Interoperability
Interoperability between software components for different stages and types of analysis is essential to the success of Bioconductor. Interoperability is established through the definition of common data structures that package authors are expected to use [18] (Table 2). Technically, Bioconductor's common data structures are implemented as classes in the S4 object-oriented system of the R language. In this manner, useful software concepts including encapsulation, abstraction of interface from implementation, polymorphism, inheritance and reflection are directly available. It allows core tasks such as matching of sample data and metadata to be adopted across disciplines, and it provides a foundation on which community development is based.
Table 2.
Key data structures for experimental and annotation data in Bioconductor.
| Container (package) | Data type |
|---|---|
| ExpressionSet (Biobase) | Matrix-like data set, where quantitative values (e.g., gene expression) are measured for many features (e.g., genes or molecules; the rows) in multiple samples (columns). This is a standard container for microarray expression data, and it is also used for other data types, e.g., drug screens. The container also stores covariates that describe the experimental factors and technical parameters associated with each feature or sample. |
| SummarizedExperiment (GenomicRanges) | Analogous to ExpressionSet; in addition, the features (rows) are associated with genomic coordinates. |
| GRanges (GenomicRanges) | Genomic coordinates and associated categorical and quantitative information, e.g., gene symbol, coverage or _p_-value. |
| VCF, VRanges (VariantAnnotation) | Extensions of the SummarizedExperiment and GRanges classes to represent variant call format. Content includes reference and alternate base content, phasing, base-call uncertainty and locus-, sample- and experiment-specific metadata [24]. |
| BSgenome (BSgenome) | Represents the genome sequence of an organism, tailored to efficient interactive manipulation with R. It can include information on conventional or user-defined mask structures and allows injection of symbols encoding sequence polymorphisms. |
It is instructive to compare such a representation to popular alternatives in bioinformatics: file-based data format conventions and primitive data structures of a language such as matrices or spreadsheet tables. With file-based formats, operations such as subsetting or data transformation can be tedious and error prone, and the serialized nature of files discourages operations that require a global view of the data. In either case, validity checking and reflection cannot rely on preformed or standardized support and need to be programmed from scratch again for every convention—or are missing altogether. As soon as the data for a project are distributed in multiple tables or files, the alignment of data records or the consistency of identifiers is precarious, and interoperability is hampered by having to manipulate disperse, loosely coordinated data collections.
3.3 Shared infrastructure for distributed development
The analysis of biological data relies on reference resources, such as genome sequences, gene models, identifiers and annotation of genes and other genomic features. Standardized R representations of these resources are provided by the project to avoid redundancy of efforts and enable data integration.
Developers also benefit from fundamental software library functions that support the operations they want to carry out. For example, the Ranges infrastructure is used directly or indirectly by 43% of all Bioconductor packages, and nearly 60% depend on Biobase and over 70% on BiocGenerics. By using shared infrastructures, developers are relieved from the task of creating and maintaining such components themselves, and they can focus on their unique domain-specific contributions.
4 Merits of a high-level language
4.1 Functionality
Software engineering is a complex process. Common expectations of scientific software include functionality, flexibility and robustness. At the early stages of developing a scientific approach, these aims should take priority, and premature optimization for speed or other hardware resources tends to be distracting. Working in a high-level language such as R is therefore a rapid and effective choice. There is plenty of time, once the right approach has been settled on, to worry about whether the computation really needs to be faster and where the bottleneck lies.
4.2 Extensibility
R provides a syntax for manipulating data. That syntax can be readily mapped to other languages. Once an idea has been vetted and tested, developers can resort to implementation of critical code sections in other languages, such as C, to improve performance. For instance, the Ranges infrastructure has gone through many iterations of this process. Alternatively, as we have done with the Rsamtools package, R's foreign language interfaces can be exploited to access an established software library from within R. This has allowed high-level code written in R to seamlessly use the functionality of the SAMtools software [19].
4.3 Reuse
Reusing software by interfacing to an existing library is one of the guiding principles of Bioconductor. Developing good software is difficult and time consuming, and if a well-tested, well-supported implementation with a suitable license already exists for a task, we encourage developers to build upon it. Using R's foreign language interfaces, they can invoke third-party software that is installed elsewhere on the system or they can include and redistribute it with their own Bioconductor package. Within the R ecosystem, the CRAN and Bioconductor repositories provide developers with access to a multitude of packages. These support rapid development because they are units that can be installed and used with little effort, and they encapsulate know-how that is often the concentrate of years of effort.
4.4 Performance and scalability
Effectively working with large data requires programming practices that match memory and processor use to available resources. R is efficient when operating on vectors or arrays, so a pattern used by high-performing and scalable algorithms is to split the data into manageable chunks and to iterate over them. An example is the yieldSize argument of functions that process BAM, FASTQ or VCF files [20]. Chunks can be evaluated in parallel to gain speed. The BiocParallel package helps developers employ parallel evaluation across different computing environments while shielding users from having to configure the technicalities. It connects to back ends for shared memory and cluster configurations. The GenomicFiles package ties parallelization to chunkwise operations across multiple files. Bioconductor is available as a virtual machine image configured for high-performance computing in Amazon's Elastic Compute Cloud (EC2).
A key aspect to the success of Bioconductor is the ability to reach both users and developers. For users, there are packages and workflows for many common use cases, as well as facilities to effectively communicate results through tables, visualization and reports. Analysis scripts are easily shared, thus facilitating reproducible research. For developers wanting to create and disseminate novel ideas, there is a well-maintained infrastructure for robust code development. Our community strives to balance user needs while simultaneously working on the leading edge of innovation in genomic data science. We are driven by the strengths and dedication of our users and developers and are optimistic about the future of the project.
Box 1: Getting started.
- Install R and Bioconductor following the directions at http://www.bioconductor.org/install. Optionally, choose an Integrated Development Environment (IDE), for example, RStudio (http://www.rstudio.com). Learn the basics of the R language, for example, with http://tryr.codeschool.com.
- Explore the Bioconductor help, http://www.bioconductor.org/help—which includes material from training courses, sample workflows, vignettes and manual pages—and the online support forum (https://support.bioconductor.org).
- Identify and install Bioconductor packages using hierarchically organized “BiocViews” and text search (http://www.bioconductor.org/packages/release/BiocViews.html), and by exploring ‘landing pages’ for package descriptions and links to vignettes, manual pages and usage statistics.
- Get to work exploring sample data sets and adapting established workflows for your own analysis!
Box 2: How to contribute?
- The first step to contributing is becoming familiar with the existing software offerings and the underlying science. Often this leads to the identification of needs for new methods or new software tools. Prospective developers should familiarize themselves with the “Developers” section of the project web page and, if they have the opportunity, attend one of the developer meetings. The project's package guidelines include those of regular (CRAN) R packages, with additional emphasis on usage-oriented documentation, sharing of common data containers, and interoperability with other packages of the project for tasks that lie up- or downstream.
- Once a package is ready for contribution, developers submit it to a package editor via the “Package Submission” web page. Feedback is given in 1–3 weeks, often with recommendations on improving the package's code, user interface or presentation. Once the package is accepted, it is added to the build server and undergoes the daily checking procedure. From then on, the package is available in the development branch of the project, and it will become part of the next release. Releases are made every 6 months, usually in April and October.
- Each package has a designated maintainer who must be responsive to an email address registered with the package. The maintainer is expected to react to errors and bugs associated with the package and typically also answers users’ requests for help with its use. When an active maintainer of a package cannot be identified, the package is orphaned and will no longer be part of subsequent releases.
- Package maintainers usually keep developing and enhancing their package even after the initial submission. Some packages have undergone impressive extensions and maturation over the years. To make their updates, maintainers access the code source via the project's version control system (Subversion; a git-bridge also exists).
Acknowledgments
We thank all contributors to the Bioconductor and R projects. Bioconductor is supported by the National Human Genome Research Institute of the US National Institutes of Health (U41HG004059 to M.M.). Additional support is from the US National Science Foundation (1247813 to M.M.) and the European Commission FP7 project RADIANT (to W.H.). A. Bruce provided graphics support for Figure 2.
Footnotes
Competing Financial Interests. The authors declare no competing financial interests.
References
- 1.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R Development Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing. 2014 [Google Scholar]
- 3.Hahne F, Huber W, Gentleman R, Falcon S. Bioconductor Case Studies. Springer; 2008. [Google Scholar]
- 4.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnishi Y, et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 2014;16:27–37. doi: 10.1038/ncb2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finak G, et al. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput. Biol. 2014;10:e1003806. doi: 10.1371/journal.pcbi.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chelaru F, Smith L, Goldstein N, Corrada Bravo H. Epiviz: interactive visual analytics for functional genomics data. Nat. Methods. 2014;11:938–940. doi: 10.1038/nmeth.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentleman R. Reproducible research: a bioinformatics case study. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1034. Article2. [DOI] [PubMed] [Google Scholar]
- 10.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat. Methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 12.Waldron L, et al. Comparative meta-analysis of prognostic gene signatures for late-stage ovarian cancer. J. Natl. Cancer Inst. 2014;106:dju049. doi: 10.1093/jnci/dju049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riester M, et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J. Natl. Cancer Inst. 2014;106:dju048. doi: 10.1093/jnci/dju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goecks J, Nekrutenko A, Taylor J, The Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez F, Granger BE. IPython: a system for interactive scientific computing. Comput. Sci. Eng. 2007;9:21–29. [Google Scholar]
- 17.Credit for code. Nat. Genet. 2014;46:1. doi: 10.1038/ng.2869. Anonymous. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S, et al. The anatomy of successful computational biology software. Nat. Biotechnol. 2013;31:894–897. doi: 10.1038/nbt.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence M, Morgan M. Scalable genomics with R and Bioconductor. Stat. Sci. 2014;29:214–226. doi: 10.1214/14-STS476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazma A, et al. Minimum information about a microarray experiment (MIAME) - toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 22.Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obenchain V, et al. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014;30:2076. doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]