Rationally engineered Cas9 nucleases with improved specificity (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 15.
Published in final edited form as: Science. 2015 Dec 1;351(6268):84–88. doi: 10.1126/science.aad5227
Abstract
The RNA-guided endonuclease Cas9 is a versatile genome editing tool with a broad range of applications from therapeutics to functional annotation of genes. Cas9 creates double-strand breaks (DSBs) at targeted genomic loci complementary to a short RNA guide. However, Cas9 can cleave off-target sites that are not fully complementary to the guide, which poses a major challenge for genome editing. Here, we use structure-guided protein engineering to improve the specificity of Streptococcus pyogenes Cas9 (SpCas9). Using targeted deep sequencing and unbiased whole-genome off-target analysis to assess Cas9-mediated DNA cleavage in human cells, we demonstrate that “enhanced specificity” SpCas9 (eSpCas9) variants reduce off-target effects and maintain robust on-target cleavage. Thus, eSpCas9 could be broadly useful for genome editing applications requiring a high level of specificity.
The RNA-guided endonuclease Cas9 from microbial CRISPR-Cas adaptive immune systems is a powerful tool for genome editing in eukaryotic cells (1, 2). However, the nuclease activity of Cas9 can be triggered even when there is imperfect complementarity between the RNA guide sequence and an off-target genomic site, particularly if mismatches are distal to the protospacer adjacent motif (PAM), a short stretch of nucleotides required for target selection (3, 4). These off-target effects pose a challenge for genome editing applications. Here, we report the structure-guided engineering of Streptococcus pyogenes Cas9 (SpCas9) to improve its DNA targeting specificity.
Several strategies to enhance Cas9 specificity have been reported, including reducing the amount of active Cas9 in the cell (3, 5, 6), using Cas9 nickase mutants to create a pair of juxtaposed single-stranded DNA nicks (7, 8), truncating the guide sequence at the 5′ end (9), and using a pair of catalytically-inactive Cas9 nucleases, each fused to a FokI nuclease domain (10, 11). Although each of these approaches reduce off-target mutagenesis, they have a number of limitations: Reducing the amount of Cas9 can decrease on-target cleavage efficiency, double nicking requires the concurrent delivery of two sgRNAs, and truncated guides can increase indel formation at some off-target loci and reduce the number of target sites in the genome (12, 13).
Cas9-mediated DNA cleavage is dependent on DNA strand separation (14, 15). Mismatches between the sgRNA and its DNA target in the first 8–12 PAM-proximal nucleotides can eliminate nuclease activity; however, this nuclease activity can be restored by introducing a DNA:DNA mismatch at that location (16–19). We hypothesized that nuclease activity is activated by strand separation and reasoned that by attenuating the helicase activity of Cas9, mismatches between the sgRNA and target DNA would be less energetically favorable, resulting in reduced cleavage activity at off-target sites (fig. S1).
The crystal structure of Streptococcus pyogenes Cas9 (SpCas9) in complex with guide RNA and target DNA (14, 15) provides a basis to improve specificity through rational engineering. The structure reveals a positively-charged groove, positioned between the HNH, RuvC, and PAM-interacting domains in SpCas9, that is likely to be involved in stabilizing the non-target strand of the target DNA (Fig. 1, A and B, and fig. S2). We hypothesized that neutralization of positively-charged residues within this non-target strand groove (nt-groove) could weaken non-target strand binding and encourage re-hybridization between the target and non-target DNA strands, thereby requiring more stringent Watson-Crick base pairing between the RNA guide and the target DNA strand.
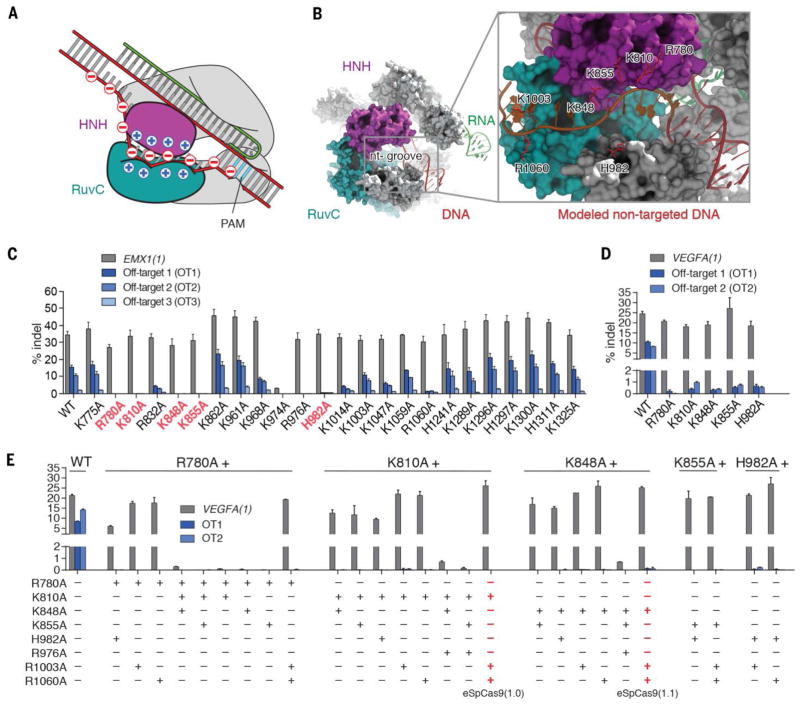
Fig. 1. Structure-guided mutagenesis improves specificity of SpCas9.
(A) A model of Cas9 unwinding highlighting locations of charge on DNA and the nt-groove. The nt-groove between the RuvC (teal) and HNH (magenta) domains stabilize DNA unwinding through non-specific DNA interactions with the non-complementary strand. RNA:cDNA and Cas9:ncDNA interactions drive DNA unwinding (top arrow) in competition against cDNA:ncDNA rehybridization (bottom arrow). (B) A crystal structure of SpCas9 (PDB ID 4UN3) showing the nt-groove situated between the HNH (magenta) and RuvC (teal) domains. The non-target DNA strand (red) was manually modeled into the nt-groove (inset). (C) Screen of alanine single mutants for improvement in specificity. The top five specificity conferring mutants are highlighted in red. (D) Assessment of top single mutants at additional off-target loci. (E) Combination mutants improve specificity compared to single mutants. eSpCas9(1.0) and eSpCas9(1.1) are highlighted in red.
To test this hypothesis, we generated SpCas9 mutants consisting of individual alanine substitutions at 32 positively-charged residues within the nt-groove and assessed changes to genome editing specificity (Fig. 1C; fig. S3, A and B; and fig. S4). Single amino acid mutants were tested for specificity by targeting them to the EMX1(1) target site in human embryonic kidney (HEK) cells using a previously validated guide sequence; indel formation was assessed at the on-target site and three known genomic off-target (OT) sites (3, 4). Five of the 32 single amino acid mutants reduced activity at all three off-target sites by at least 10-fold compared to wild-type (WT) SpCas9 while maintaining on-target cleavage efficiency, and 6 others improved specificity 2 to 5-fold. These mutants also exhibited improved specificity when tested on a second locus, VEGFA(1) (Fig. 1D).
Although some single amino acid mutants were more specific than WT SpCas9 when targeting EMX1(1) and VEGFA(1), off-target indels were still detectable (~0.5%) (Fig. 1D). To further improve specificity, we performed combinatorial mutagenesis using the top single amino acid mutants identified in the initial screen. Eight out of 35 combination mutants retained wild-type on-target activity and displayed undetectable off-target indel levels at EMX1(1) OT1, VEGFA(1) OT1, and VEGFA(2) OT2 (Fig. 1E and fig. S3, C and D).
To ensure that the observed decrease in off-target activity was not accompanied by reduced on-target activity, we measured on-target indel formation at 10 target sites in 3 genomic loci using the top 14 mutants (fig. S5) and ranked these based on a combination of preserved on-target activity and decreased off-target activity. We identified three mutants with both high efficiency (wild type levels of on-target indel formation) and specificity (no detectable indel formation at EMX(1) and VEGFA(1) off-targets): SpCas9 (K855A), SpCas9 (K810A/K1003A/R1060A) [also referred to as eSpCas9(1.0)], and SpCas9 (K848A/K1003A/R1060A) [also referred to as eSpCas9(1.1)]. These three variants were selected for further analysis.
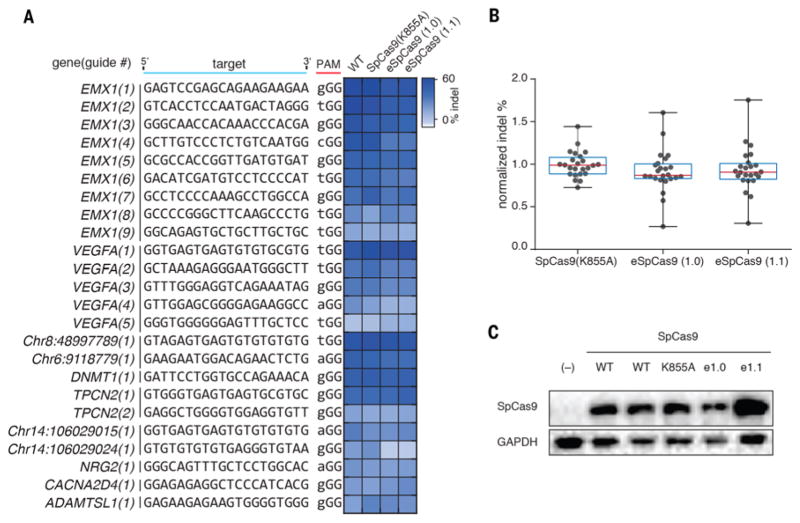
We expanded this assay to assess whether SpCas9 (K855A), eSpCas9(1.0), and eSpCas9(1.1) broadly retained efficient nuclease activity, measuring on-target indel generation at 24 target sites spanning 10 genomic loci (Fig. 2A). All three mutants generated similar indel levels as WT SpCas9 with the majority of target sites (Fig. 2B). Mutants were expressed equivalently or at higher levels than WT SpCas9 based on a Western blot (Fig. 2C), indicating that improvements in specificity were not due to decreased protein expression levels.
Fig. 2. SpCas9 mutants maintain on-target efficiency.
(A) Assessment of mutants for efficient on-target cutting with 24 sgRNAs targeted to 10 genomic loci. (B) Tukey plot of normalized on-target indel formation for mutants. (C) Western blot of SpCas9 using anti-SpCas9 antibody.
We compared the specificity of the three mutants to WT SpCas9 with truncated guide sequences [18 nt for EMX1(1) and 17 nt for VEGFA(1)], which have been shown to reduce off-target indel formation (12) (fig. S6). When using full-length (20 nt) guides, all three mutants reduced cleavage at all off-target sites assessed. Specifically, eSpCas9(1.0) and eSpCas9(1.1) with 20 nt RNA guides significantly reduced or eliminated cleavage at 22 out of 24 off-target sites (< 0.2% indel). In contrast, WT SpCas9 with truncated guides (17 nt for VEGFA3 or 18 nt for EMX1) eliminated 14 of 24 sites but also increased off-target activity at 5 sites compared to WT SpCas9 with 20 nt guides.
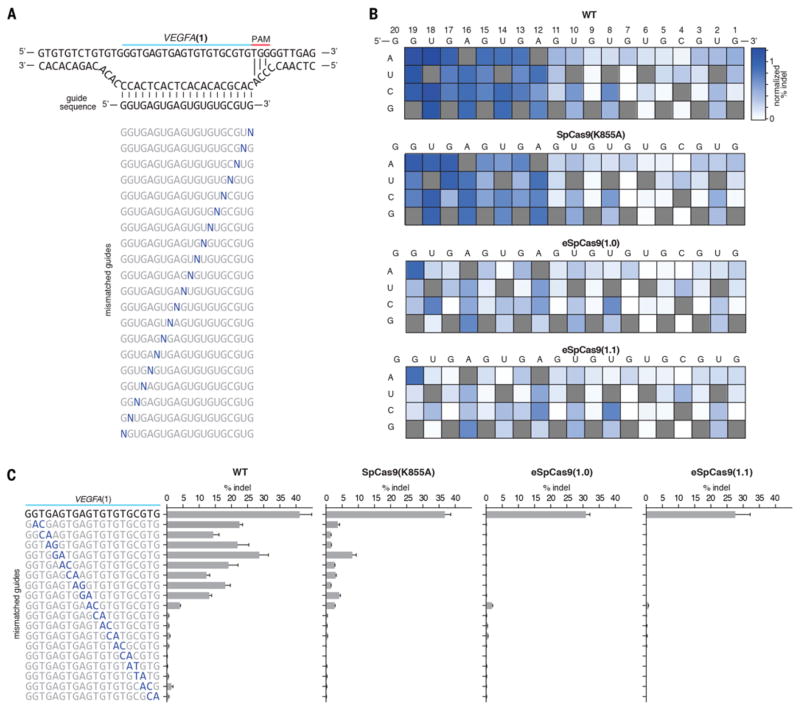
To further understand the tolerance SpCas9 (K855A), eSpCas9(1.0), and eSpCas9(1.1) for mismatched target sites, we systematically mutated the VEGFA(1) guide sequence to introduce single and double base mismatches at different positions (Fig. 3, A to C). Compared to WT SpCas9, all three mutants induced lower levels of indels with mismatched guides. Of note, eSpCas9(1.0) and eSpCas9(1.1) induced lower indel levels even with single base mismatches located outside of the 7–12bp seed sequence. Given that we did not observe any difference between eSpCas9(1.0) and eS-pCas9(1.1) in terms of specificity, we selected SpCas9 (K855A) and eSpCas9(1.1) for further analysis based on on-target efficiency.
Fig. 3. SpCas9 mutants exhibited increased sensitivity to single and double base mismatches between the guide RNA and target DNA.
(A) Schematic showing design of mismatched guide sequences against VEGFA(1). (B) Heat maps showing indel % of guide sequences with a single base mismatch. (C) Indel formation with guide sequences containing consecutive transversion mismatches.
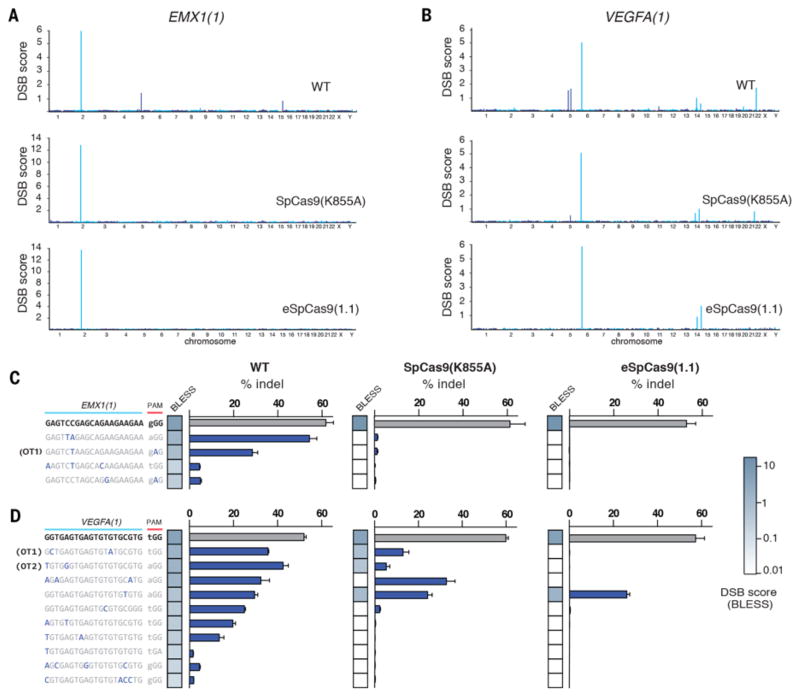
We assessed the genome-wide editing specificity of SpCas9 (K855A) and eSpCas9(1.1) using BLESS (direct in situ breaks labeling, enrichment on streptavidin and next-generation sequencing) (20, 21), which quantifies DNA double-stranded breaks (DSBs) across the genome (fig. S7A), for both the EMX1(1) and VEGFA(1) targets for both mutants and compared these results to WT SpCas9. We used a previously established computational pipeline for distinguishing Cas9 induced DSBs from background DSBs (21) (fig. S7B). Both SpCas9 (K855A) and eSpCas9(1.1) exhibited a genome-wide reduction in off-target cleavage and did not generate any new off-target sites (Fig. 4, A to D).
Fig. 4. Unbiased genome-wide off-target profile of mutants using BLESS.
(A and B) Manhattan plots of genome-wide DSB clusters generated by each SpCas9 mutant using the EMX1(1) and VEGFA(1) targeting guides. (C and D) Targeted deep sequencing validation of off-target sites identified in BLESS. Off-target sites are ordered by DSB score (blue heatmap). Green heatmaps indicates sequence similarity between target and off-target sequences.
These findings also provide insight into the mechanism of Cas9 targeting and nuclease activity. We propose that off-target cutting occurs when the strength of Cas9 binding to the non-target DNA strand exceeds forces of DNA re-hybridization. Consistent with this model, mutations designed to weaken interactions between Cas9 and the non-complementary DNA strand led to a substantial improvement in specificity. The model also suggests that, conversely, specificity can be decreased by strengthening the interactions between Cas9 and the non-target strand. Consistent with this hypothesis, we generated two mutants, S845K and L847R, each of which exhibited decreased specificity (fig. S8). Similar strategies described in this study can also be successfully applied to other Cas9 family proteins such as Staphylococcus aureus Cas9 (SaCas9) (fig. S9) to engineer nucleases with improved specificity.
In conclusion, we have demonstrated through structure-guided design that neutralization of positive charges in the nt-groove can dramatically decrease off-target indel formation while preserving on-target activity. These data show that eSpCas9(1.1) can be used to increase the specificity of genome editing applications. Future structure-guided interrogation of Cas9 binding and cleavage mechanism will likely enable further optimization of the CRISPR-Cas9 genome editing toolbox.
Supplementary Material
SUPP
Acknowledgments
We thank J. Dahlman for helpful discussions and a critical review of the manuscript; F. A. Ran, R. J. Platt, and J. Joung for experimental assistance; and the entire Zhang laboratory for support and advice. I.S. is supported by the Simons Center for the Social Brain, F.Z. is supported by the National Institutes of Health through NIMH (5DP1-MH100706) and NIDDK (5R01DK097768-03), a Waterman Award from the National Science Foundation, the Keck, New York Stem Cell, Damon Runyon, Searle Scholars, Merkin, and Vallee Foundations, and B. Metcalfe. F.Z. is a New York Stem Cell Foundation Robertson Investigator. I.S., L.G., B.Z., and F.Z. are inventors on provisional patent application 62/181,453 applied for by the Broad Institute and MIT that covers the engineered CRISPR proteins described in this manuscript. Plasmid DNA encoding eSpCas9(1.0) and eSpCas9(1.1) are available from Addgene under a Universal Biological Material Transfer Agreement with the Broad Institute and MIT. F.Z. is a founder and scientific advisor for Editas Medicine and a scientific advisor for Horizon Discovery. Further information about the protocols, plasmids, and reagents can be found at the Zhang laboratory website (www.genome-engineering.org).
Footnotes
REFERENCES AND NOTES
- 1.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Reyon D, Joung JK. Targeted genome editing in human cells using CRISPR/Cas nucleases and truncated guide RNAs. Methods Enzymol. 2014;546:21–45. doi: 10.1016/B978-0-12-801185-0.00002-7. [DOI] [PubMed] [Google Scholar]
- 10.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJR, Boekema EJ, Dickman MJ, Doudna JA. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, Pasero P, Rowicka M, Dikic I. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPP