A global profile of replicative polymerase usage (original) (raw)
. Author manuscript; available in PMC: 2016 Mar 14.
Published in final edited form as: Nat Struct Mol Biol. 2015 Feb 9;22(3):192–198. doi: 10.1038/nsmb.2962
Abstract
Three eukaryotic DNA polymerases are essential for genome replication. Polα-primase initiates each synthesis event and is rapidly replaced by processive DNA polymerases: Polε replicates the leading strand while Polδ performs lagging strand synthesis. However, it is not known whether this division of labour is maintained across the whole genome or how uniform it is within single replicons. Using S. pombe, we have developed a polymerase usage sequencing (Pu-seq) strategy to map polymerase usage genome–wide. Pu–seq provides direct replication origin location and efficiency data and indirect estimates of replication timing. We confirm that the division of labour is broadly maintained across an entire genome. However, our data suggest a subtle variability in the usage of the two polymerases within individual replicons. We propose this results from occasional leading strand initiation by Polδ followed by exchange for Polε.
Introduction
Accurate DNA replication is fundamental to life and errors that occur during replication underpin the genome instability that is the hallmark of cancer development1,2. In most eukaryotes, bidirectional replication is initiated stochastically, with distinct regions of the genome showing varying initiation efficiencies and distinct temporal regulation3. In budding yeast, specific DNA consensus sequences define the binding of the origin recognition complex (ORC) to DNA throughout the cell cycle4. Each region of replication initiation is thus defined by a single DNA sequence or origin. In higher eukaryotes ORC association with the chromosomes varies through the cell cycle and the mechanisms defining where ORC binds are not understood. Initiation zones in higher eukaryotes are likely composed of numerous low efficiency origins clustered together3.
In exponentially growing budding yeast the different origins are activated with different efficiencies. Thus, times at which different initiation regions are replicated (the population average) are distinct5. In higher eukaryotes, growing cultures of individual cell types display reproducible replication timing profiles indicating that ORC association and or the likelihood of replication initiation from ORC associated regions are stable characteristics of specific cell types6. Interestingly, timing profiles for different mammalian cell types correlate well with 3-D chromosome interaction maps, suggesting a link between replication timing and chromatin organisation within the nucleus (reviewed in:3).
ORC attracts the MCM complex in G1 phase of the cell cycle, licensing the site for initiation7. The six subunit MCM complex is the core of the replicative helicase, which is subsequently activated by the loading of two additional components; Cdc45 and the four subunit GINS complex. The resulting active helicase is known as CMG8. An ancillary replisome component, the Ctf4 trimer, links Polα–primase to CMG9,10, coordinating the necessary initiation events. The Polε holoenzyme interacts directly with GINS, an association also required independently for the initial formation of CMG11-13. Once CMG is formed, the Polε holoenzyme–GINS interaction is not required for CMG helicase activity. It is not known if the Polδ holoenzyme interacts directly with CMG. Once DNA replication is initiated, each fork synthesises the leading strand continuously and the lagging strand discontinuously.
Certain DNA polymerase mutations introduce a biased mutation spectrum. This has allowed assignment of the source of mutations to mispairing on one or the other DNA strand14. Using these mutant polymerases, Polε was genetically assigned as the leading strand DNA polymerase at several loci in S. cerevisiae. Similarly, Polδ was assigned as the major lagging strand polymerase15. These data led to the model that the labour of replication is shared: Polε replicates the leading stand and Polδ the lagging strand. An equivalent experiment using _S. pomb_e similarly assigned Polδ to the lagging strand16, demonstrating evolutionary conservation of polymerase usage. An S. pombe mutant Polε that incorporated ribonucleotides into DNA at increased frequency was used to physically assign Polε to leading strand synthesis16. These experiments relied on the increased incorporation of rNTPs into the leading strand, causing that specific stand to be fragmented by alkali treatment, which cleaves the phosphate backbone at ribonucleotides but not deoxyriboncleotides.
To establish if the division of labour between Polε and Polδ is consistent across an entire genome and to ascertain if there is variation in the usage between the two polymerases within a single replicon we set out to physically map, genome–wide, the division of labour between these polymerases. We devised a strategy to identify, by high throughput sequencing, the position of ribonucleotides in the genome and combined this with Polε and Polδ mutants that incorporate excess ribonucleotides to establish a polymerase usage sequencing (Pu–seq) methodology that allowed us to map the division of labour genome–wide. We confirm that the division of labour is broadly maintained across an entire genome. We also demonstrate that a single Pu–seq experiment, which consists of two library samples for deep sequencing (one each from asynchronous cultures of the respective polymerase mutants) delivers a direct and extremely high resolution genome–wide map of DNA replication initiation and allows the indirect calculation of robust genome–wide replication timing data. The resolution of our data revealed evidence for subtle variability in the usage of the two polymerases within individual replicons. We suggest this results from occasional leading strand initiation by Polδ.
Results
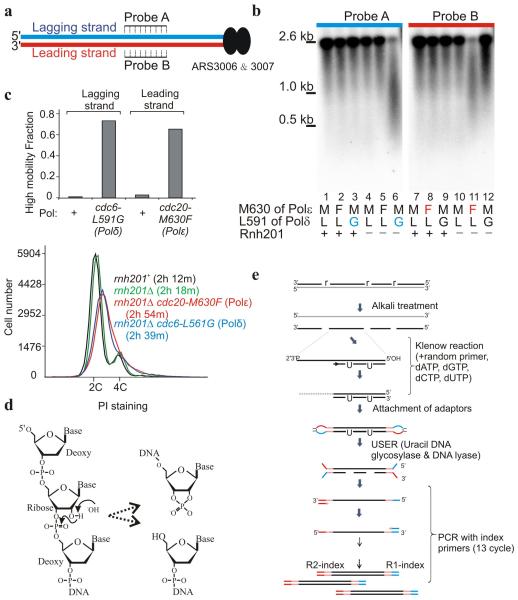
At physiological dNTP and rNTP concentrations S. cerevisiae replicative DNA polymerases incorporate, in vitro, ribonucleotides at frequencies ranging from 1:650 bp (Polα) to 1:5000 bp (Polδ)17. Ribonucleotides are efficiently removed from duplex DNA by ribonucleotide excision repair (RER): RNAseH2 nicks 5′ to the ribonucleotide, Polδ (or Polε) initiates strand–displacement synthesis and Fen1 (or Exo1) removes the resulting flap before ligation completes repair18. In the absence of RER single ribonucleotides persist (although some are removed by Topo119-21). Ribonucleotides can template DNA synthesis, albeit with a reduction in processivity22,23. We previously exploited an S. pombe cdc20–M630F (Polε) allele to introduce excess ribonucleotides into DNA replicated by Polε. Southern blot analysis in an RnaseH2–deficient (_rnh201_Δ) background provided physical evidence that Polε performed the majority of leading strand synthesis16. To facilitate mapping the division of labour genome–wide, we have generated an equivalent mutation for Polδ, cdc6–L591G. DNA prepared from cells harbouring this mutation showed lagging strand–specific degradation when alkali gels were probed for sequences flanking an efficient origin (Fig. 1a,b). This is complementary to the DNA prepared from cells harbouring the previously characterised cdc20–M630F (Polε) allele, which demonstrated leading strand–specific degradation (Fig. 1b). Both the cdc20–M630F (Polε) and the cdc6–L591G (Polδ) mutant strains in the _rnh201_Δ background incorporated similar levels of ribonucleotides24, grew with similar kinetics and displayed similar flow cytometry profiles (Fig. 1c).
Figure 1.
rNMP incorporation into DNA in Polδ (cdc6–L591G) and Polε (cdc20–M630F) cells. (a) Schematic representation of the region flanking ARS3006 and 3007. Leading and lagging strands are represented by red and blue lines, respectively. (b) Southern blot of digested and alkali treated genomic DNA hybridized with probes indicated in panel a. (c) Top: the proportion of high mobility product from _rnh201_Δ cells in experiments equivalent to panel b. Bottom: flow cytometry analysis of wild type, _rnh201_Δ, _rnh201_Δ cdc20 –M630F (Polε) and _rnh201_Δ cdc6–L591G (Polδ) cells with population doubling times in parenthesis. (d) Hydrolysis at the misincorporated RNA molecule. The 2′ OH group of the rNMP is susceptible to nucleophilic attack (left), causing cleavage of the sugar backbone and the generation of a cyclic 2′3′ phosphate and a 5′ OH group. (e) Schematic of library preparation. Position of incorporated ribonucleotides shown as “r”.
Mapping polymerase usage across the genome
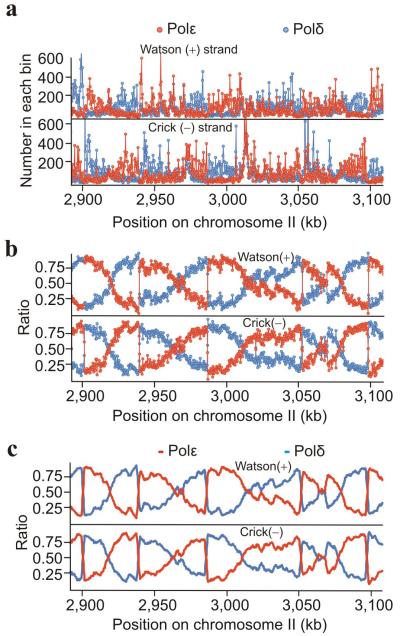
Alkali treatment of duplex ribonucleotide–containing DNA results in phosphate backbone cleavage 3′ to the ribose resulting in a 5′OH (Fig. 1d). If the denatured DNA is used to template random hexamer primer extension, 5′ to 3′ synthesis results in a flush end adjacent to the initial ribose (Fig. 1e). By generating a library from single–stranded DNA and placing distinct index primers at each end, deep sequence reads can be mapped to individual strands, locating with base accuracy the original ribonucleotide. To map replication polymerase usage across the genome we therefore grew two RnaseH2–deficient cultures harbouring cdc20–M630F (Polε) or polδ–L591G (Polδ) mutations, prepared DNA, treated this with alkali and created two independent libraries. Approximately 10 million paired–end sequence reads for each strain were mapped to 300 bp bins across the genome (Fig. 2a). The relative ratio of reads from the Polε and Polδ datasets was calculated (Fig. 2b) and the data smoothed to provide frequency scores representative of relative Polε and Polδ usage for the Watson (+) and Crick (−) strands (Fig. 2c).
Figure 2.
Polymerase usage across the fission yeast genome. (a) Total counts of the flanking 5′ nucleotide of the sequenced reads assigned to 300 bp bins plotted for a representative region (Polε (cdc20–M630F; red), Polδ (cdc6–L519G; blue). (b) Ratio of the relative reads in each bin for Polε (cdc20–M630F: (ε/[ε+δ], red) and Polδ (cdc6–L519G: (δ/[δ+ε], blue) plotted for the same region. (c) Smoothed data providing a map of polymerase usage (see also supplementary Fig. 2). Supplementary datasets to visualise the whole genome are listed in supplementary Table 2.
Polymerase usage transitions define initiation sites
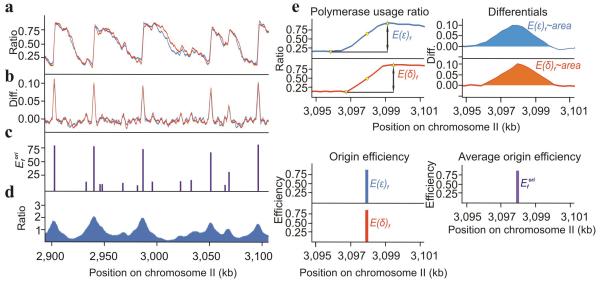
Bidirectional initiation and the division of polymerase labour predicts a reciprocal demarcation on both the Watson and the Crick strands between Polε (leading) and Polδ (lagging) usage for each initiation zone. Efficient origins should manifest as sharp reciprocal changes in the polymerase usage ratios. Less efficient origins, which are replicated passively in most cells, should present as reciprocal inflections in otherwise uniform gradients. The two independent datasets were thus used to calculate Polε usage on the Watson stand or Polδ usage on the Crick strand (Fig. 3a) and the differential of each neighbouring data point plotted (Fig. 3b). Where a reciprocal positive peak was identified (i.e. change in polymerase usage in both data sets), maxima and minima were derived (Fig. 3c) and the average of their differences plotted (Fig. 3d). Peak heights reflect relative origin efficiency: the highest peaks correspond to the most efficient origins.
Figure 3.
Identification of replication origins. (a) The usage of Polε on the Watson (blue) and Polδ on the Crick (red) strand. (b) The differential (Diff.) of the polymerase usage plots from panel a. (c) Origin efficiencies (Efori) calculated from Pu–seq data. (d) A comparative map of origins generated by BrdU IP–seq from YD18 cells synchronised by cdc25 (G2) block and release into HU (see also supplementary Fig. 2). Supplementary datasets to visualise the whole genome are listed in supplementary Table 2. (e) Example of how origin efficiencies were quantified. Top left: Established minima and maxima (yellow triangles) around the reciprocal peaks (yellow dots) identified from panel b. Top right: example region of differentials from panel b. Bottom left: Differences between the above identified maxima and minima (E(δ)f and E(ε)f). Bottom right: Averaged differences producing the relative origin efficiency (Efori).
The distribution of origin efficiencies is given in supplementary Fig. 1a. The origins identified and their relationship to previous studies are presented in supplementary Table1. To account for experimental variation we analysed four additional independent experiments and annotated how often each origin was identified (supplementary Table1). To independently visualise origins in a manner coherent with the literature25, we synchronised wild–type cells in G2, released them into S phase in the presence of hydroxyurea (HU) plus the nucleotide analogue bromo–deoxyuridine (BrdU) and quantified replication using BrdU immunoprecipitation plus deep sequencing (Fig. 3e). This identified 421 origins, >90% of which correspond to Pu–seq origins (supplementary Table 1 and supplementary Fig. 2).
A map of replication timing by marker frequency analysis
While Pu–seq provides a direct assay for replication initiation efficiency, it can also indirectly provide information about relative replication timing (see below). To validate replication timing data calculated from the Pu–seq experiments, we first wished to generate a direct replication timing map for S. pombe that is not biased by cell synchronisation or treatment with replication inhibitors25. We thus mapped replication profiles of cells synchronised by elutriation using marker frequency analysis (Fig. 4a). Aliquots of an elutriated culture were examined over time for mitotic index, septation and DNA content. Based on the known cell cycle behaviour of S. pombe, these data were used to calculate percentages of G2, mitotic, S phase and post S phase cells for each time point (Fig. 4b, see Materials and Methods). The fraction of DNA replicated for each time point was then calculated and boundaries set for the beginning and end of S phase (Fig. 4c). DNA from the indicated aliquots spanning S phase was extracted and libraries prepared for deep sequencing. The proportion of reads for each 1kb bin across the genome was compared to a fully replicated (G2) control and the percentage of replication calculated at each locus for each time point sequenced (Fig. 4d).
Figure 4.
Genome replication timing in S. pombe. (a) Flow cytometry profiles of cells synchronised in G2 by elutriation, washed into fresh media and allowed to progress through mitosis and into S phase. (b) The percentage of cells in G2, mitosis, S phase and post S phase cells. (c) The population–average genome copy number calculated for each time point. The period in which cells in the population are in S phase is shaded blue. (d) Visualisation of DNA copy number during the S phase time course across a representative region. Open circles define origins.
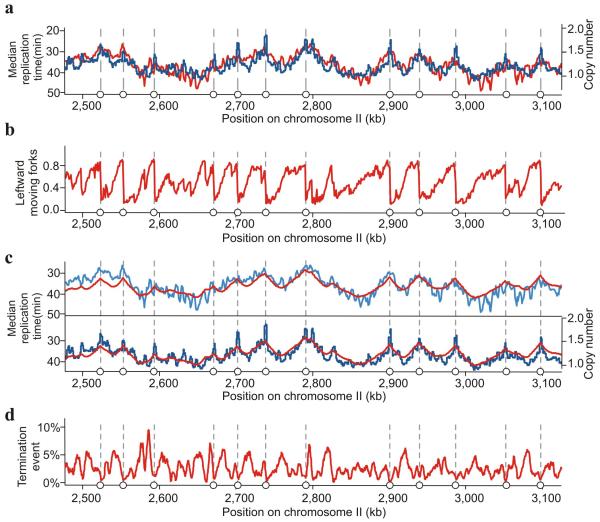
Because elutriation can cause cellular perturbation due to centrifugation26 we validated that elutriation did not distort replication profiles by performing sort–seq analysis: S phase cells are recovered by fluorescence activated cell sorting from an asynchronous culture and subjected to deep sequencing27. Plotting the normalised copy number for the sort–seq against the calculated median replication time from the marker frequency analysis of the elutriated culture demonstrated a good correlation (Fig. 5a). This confirms that elutriation does not perturb replication timing.
Figure 5.
Characterisation of DNA replication profiles. (a) Comparison of Trep (median replication time – the time at which 50% of the locus is replicated) calculated from the synchronous culture by marker frequency analysis (see Fig. 4d; red) and the normalised copy number of each locus from a single population of cells sorted by FACS from an asynchronous culture (sort–seq; blue). Open circles define origins. (b) The percentage of leftward moving forks calculated from the Pu–seq data. (c) Comparison of Trep derived from marker frequency analysis and sort–seq with Trep determined by Pu–seq. In both panels the red line represents Trep calculated from the Pu–seq data. In the top panel, the blue line represents Trep calculated from the marker frequency analysis. In the bottom panel, the blue line shows the copy numbers derived from sort–seq. (d) The calculated percentage of replication termination events from the Pu–seq data for each locus. Supplementary datasets to visualise the whole genome are listed in supplementary Table 2.
Pu–Seq provides timing and termination information
Mathematical analysis of the Pu–seq provides a measure of replication timing: the proportion of reads mapping to each strand from the cdc6–L519G (Polδ) and cdc20–M630F (Polε) datasets provides two independent and direct measurements of the proportion of replication forks moving leftward (or rightward) throughout the genome (Fig. 5b). Such fork direction data allows a direct calculation of relative replication times5,28. Based upon a mean replication fork velocity of 1.5 kb/min we calculated a relative replication timing map from Pu–seq data that is superimposable on direct replication time measurements derived from the time–course and sort–seq analysis (Fig. 5c). Changes in mean fork direction across a chromosome are a consequence both of replication origin activity and of replication termination events: even close to an efficient origin, the proportion of moving forks always decreases with distance. This is the consequence of both the initiation and replication termination events in the population. We can thus also calculate the percentage of termination events occurring within a defined window. While we observe that replication origins result in sharp transitions in fork direction, indicating discrete and efficient initiation sites, replication termination events are dispersed stochastically across large termination zones (Fig. 5d) with no evidence of programmed termination regions (supplementary Fig. 1b).
Observed polymerase usage variation within a replicon
Potential differences in the ribonucleotide incorporation rates between cdc20–M630F (Polε) and cdc6–L519G (Polδ) preclude establishing accurately the absolute fraction of DNA synthesised by Polε and Polδ. Without considering the minor contribution of Polα, the anticipated division of labour and coupled leading plus lagging strand synthesis predicts ~50% of the genome is replicated by Polε and ~50% by Polδ. Using this assumption, we plotted polymerase usage of the duplex for each 300 bp bin across the genome (Fig. 6a). Genome–wide, the division of labour was largely uniform, although small fluctuations are evident. The majority of these correspond to efficient origins. Therefore, we computationally identified inter–origin regions of >30 kb where the directionality of replication forks was not appreciably perturbed by less efficient origins (Fig. 6b) and determined the average use of Polε and Polδ across replicons. A significant bias towards Polδ was evident proximal to origins, which declined towards the centre of the inter–origin region. This effect was not influenced by either global replication timing or by the absence of the Rad18 ubiquitin ligase (supplementary Fig. 1c), which prevents PCNA ubiquitination and thus compromises non–canonical polymerase usage. Thus, proximal to efficient origins, replicons exhibit an apparent bias towards Polδ usage relative to Polε that is dependent on distance from the origin and independent of post–replication repair.
Figure 6.
Asymmetric polymerase usage within a replicon. (a) Two example regions showing polymerase usage across inter–origin regions. Top panels: the ratios of usage of Polε (red) and Polδ (blue) on the Watson Strand and Crick strand. Bottom panels: total polymerase usage on duplex DNA. (b) Top: the 85 inter–origin regions between high efficiency origins (Efori > 40%) of >30 kb which do not contain lower efficiency origins (20% < Efori < 40%) are displayed as a heat map aligned to the 3 chromosomes (right bar). Light pink represents early replicating regions, brown represents late replicating regions (see supplementary Fig. 1c). Each row represents an inter–origin region. The horizontal axis shows the relative position between origins. Bottom: average values. SD = standard deviation. Supplementary datasets to visualise the whole genome are listed in supplementary Table 2.
Discussion
We have developed an approach to identify the genome–wide location of ribonucleotides incorporated into DNA. In a cdc20+ cdc6+ (Polε+ and Polδ+) _rnh2_Δ background we observe that the percentage of each ribonucleotide incorporated shows little bias compared to genomic sequence composition (supplementary Fig. 3). This is not appreciably altered in the two polymerase mutant backgrounds. We observed a moderate increase in the frequency of ribonucleotide incorporation in gene coding regions when compared to 5′ and 3′ untranslated regions and promoters, a bias that is not influenced by our polymerase mutations (supplementary Fig. 4). Adaptations to this hydrolysis dependent ribonucleotide mapping methodology will facilitate research into the causes of, and biological consequences arising from, ribonucleotide incorporation.
To study DNA replication, we combined this approach with ribonucleotide discrimination mutations in the two main replicative polymerases14-16 to provide a polymerase usage sequencing (Pu–seq) strategy that allowed us to map polymerase usage genome–wide. Our analysis demonstrated that the division of labour for Polε and Polδ is consistent across an entire genome. While not unexpected, this is important to establish. Strikingly, Pu–seq provided a highly discriminatory dataset that directly revealed the location and efficiency of replication origins at very high resolution. We compared our origin assignments to those previously collated from the literature in oriDB29. To locate potential overlap, we first identified the central nucleotide of the Pu–seq identified origin and established if it fell within plus or minus 900 bp of the reported origin region. Comparing the two datasets (741 origins from oriDB and 1145 origins recognised by Pu–seq), 97.5% of “confirmed”, 84.9% of “likely” and 67.7% of “dubious” oriDB origins were identified (supplementary Table1).
Previous work in S. cerevisiae used replication timing data to calculate termination frequencies across the genome5 and demonstrated that defined termination zones were not common: termination events per 1 kb fluctuated between approximately 0 and 4% per cell cycle across the genome. Applying this established mathematical analysis5,28 to the Pu–seq data similarly predicted that the distribution of termination frequencies in S. pombe is consistent with there not being defined termination zones between origins. This suggests that termination is largely defined by stochastic origin usage5 as opposed to the positioning of discrete replication fork pausing elements30.
The high definition provided by Pu–seq enabled us to identify an apparent bias towards Polδ close to the sites of efficient initiation, a phenomenon that is reproducible across multiple biological and experimental replicates (data not shown). This phenomenon is not influenced by either regional replication timing31 or by post–replication repair32, implying it is independent of non–canonical repair polymerases. While we cannot exclude an unidentified prosaic explanation accounting for these data, one interpretation is that a small fraction of leading strand replication events, once started by Polα–primase, are initially extended by Polδ in place of Polε.
The interaction between the N–terminal 103 amino acids of the Dpb2 subunit of Polε and GINS is likely to position Polε for leading strand synthesis. Despite the fact that this same interaction is required for the formation of the CMG complex11,13, it is subsequently dispensable for CMG helicase activity and loss of the interaction does not prevent replication progression if CMG formation is promoted by an ectopically expressing N–terminal region of Dpb111. In such cells replication is slow and synthesis of the leading strand is probably completed by Polδ. Indeed, in yeasts, the entire genome can be replicated without the catalytic activity of Polε11,33,34, demonstrating substantial flexibility in the use of Polε and Polδ during DNA replication.
The choice of Polε for leading strand synthesis is, in part at least, a function of the interaction of Polε holoenzyme and the core replication machinery discussed above. Polδ, while not apparently showing a strong interaction with the core replisome, does have a high affinity for PCNA and therefore potentially could compete for the leading strand primer. Initiation of leading strand synthesis by Polδ is likely to result in Polδ being subsequently displaced by Polε during elongation. Indeed, in vitro studies show that S. cerevisiae Polε holoenzyme is preferentially recruited to leading strand substrates pre–loaded with CMG and that, while Polδ can load in the absence of Polε, it is displaced if Polε is added after DNA synthesis has initiated35. We thus propose that the apparent discrepancy in polymerase usage within a replicon reflects occasional recruitment of Polδ to leading strand synthesis, with its subsequent displacement during progression by Polε. It will be interesting to test this proposition with further experiments in the future.
In summary, Pu–seq provides a simple, yet powerful, tool to explore genome replication in any eukaryote where suitable polymerase mutants can be introduced in a background deficient (or depleted) for RnaseH2. Unlike replication timing data, Pu–seq data directly identifies regions of replication initiation. We show here that it can also provide indirect, but accurate, evidence of relative replication timing and the frequency of termination. Pu–seq will thus provide a useful tool for examining DNA replication.
Online Methods
Genetics and mutation
Standard S. pombe genetic and molecular techniques were employed as described previously36. The cdc6–L591G (Polδ) mutant was constructed by site–directed mutagenesis and introduced into S. pombe genome by recombination–mediated cassette exchange (RMCE)37. Southern blot to detect alkali–sensitive sites in genomic DNA was performed as described previously16. A list of strains used is given in supplementary Table3.
Identification of a Polδ mutant that incorporates rNTPs
DNA containing ribonucleotides is alkali labile38, which causes strand fragmentation following alkali treatment. Exploiting an rnh201 null mutation (where RNAseH2 activity is missing) alkali–degradation and Southern blot analysis16 we assessed a range of polδ alleles with mutations of the steric gate residue L591 for their ability to incorporate ribonucleotides. cdc6–L591G (Polδ) was selected because it caused strand–specific alkali sensitivity and showed no obvious cellular phenotype. The uncropped Southern blot used in Fig. 1b is shown as Supplementary Data Set 1 in supplementary Fig. 5.
Library production and sequencing
Cells from early log phase IM642, IM855, IM654, YAK139 and YAK138 were harvested by centrifugation and genomic DNA was prepared using the QIAGEN 100/G Genomic–tip. For Pu–Seq analysis, 20 μg of genomic DNA was alkaline treated in 0.23 M NaOH at 55 °C for 2 hours. 10 μg of the single stranded DNA (ssDNA) was loaded onto 2% TBE gel and was run for 2h at 100V. The gel was stained with acridine orange (final conc. 5 μg/ml) for 2h at room temperature with gentle shaking followed by overnight destaining in water. Fragments of 300–500 bp were excised from the gel and isolated using a gel extraction kit (MACHEREY–NAGEL, NucleoSpin® Gel and PCR Clean–up).The experimental design for strand–directed high–throughput DNA sequencing was adapted from Zhang et al, (2012)39: 100 ng of purified ssDNA fragments were converted to dsDNA, using the BioPrime DNA Labelling system (invitrogen) according to manufacturer’s instructions with dNTP’s in which dTTP was substituted by dUTP. Converted dsDNA was purified by AMpure XP beads (Beckman Coulter, Inc.), concentration was determined by spectrometry (Pico green; Life Technologies) and size distribution examined using an Agilent bioanalyzer. All DNA (20 – 60 ng) was used for Illumina library preparation using NEBNext® Ultra™ DNA Library Prep Kit with the following modified protocol: end–cleaning of DNA fragments and adaptor–ligation was performed as instructed by the manufacturer but without USER treatment and followed by size selection of insert (250 – 600 bp) using AMpure XP beads. Purified DNA was then treated with USER enzyme and subjected to subsequent PCR (13 cycles) using multiplexing index–primers to generate Illumina libraries. After purification with AMpure XP beads, libraries were subjected to 100 or 150 bp paired–end sequencing using an Illumina Hiseq2500 or NextSeq 500 platform, respectively.
Analysis of Polymerase usage
Paired–end reads of high throughput sequencing were aligned to the S. pombe genome sequence (ASM294v2.23: chromosomes I, II and III, downloaded from ‘PomBase’ website) using bowtie2–2.2.2. Using alignment data, the position of the 5′ end of each R1 read, which corresponds to 5′-end of ssDNA hydrolysed by alkaline treatment, was determined and the number of reads in 300 bp bins across genome were counted separately for the Watson and Crick strands. This generated the four datasets: at the chromosome coordinate x, N+δ(x) – the count for cdc6–L591G (Polδ) on Watson strand, N_−_δ(x) – cdc6_–_L591G (Polδ) for the Crick strand, N+ε(x) – cdc20_–_M630F (Polε) for the Watson strand, N_−_ε(x) – cdc20_–_M630F (Polε) for the Crick strand. The datasets were normalised with the total number of reads: D(x)+ = N+δ(x)/ΣN+δ – Polδ mutant for Watson strand, D(x)− = N−δ(x)/ΣN−δ – Polδ – mutant for Crick strand, E(x)+ = N+ε(x)/ΣN+ε –Polε mutant for Watson strand, E(x)− = N−ε(x)/ΣN−ε –Polε mutant for Crick strand. Making the assumption that each part of the duplex genome is replicated by Polδ and Polε, the ratio of DNA synthesis catalysed by Polδ (D’) and Polε (E’) were calculated: D’(x)+ = D(x)+/(D(x)+ +E(x)+) The ratio of Polδ–synthesis on Watson strand, E’(x)+ = E(x)+/(D(x)+ + E(x)+) of Polε–synthesis on Watson strand, D’(x)− = D(x)−/(D(x)− + E(x)−) of Polδ–synthesis on Click strand, E’(x)− = E(x)−/(D(x)− +E(x)−) of Polε–synthesis on Crick strand. Using the assumption that 50% of the genome is replicated by Polε and 50% by Polδ, the ratios of Pol–usage were optimised: when n is the total number of bins, D”(x)+ = D’(x)+ × 0.5 × n/Σ D’+, D”(x)− = D’(x)− × 0.5 × n/Σ D’−, E”(x)+ = E’(x)+ × 0.5 × n/Σ E’+, E”(x)− = E’(x)− × 0.5 × n/Σ E’−. The total usage of each polymerase on both strands (Watson and Click strand) was calculated: D(x)& = (D”(x)+ + D”(x)−)/2 the ratio of Polδ-synthesis on both strands, E(x)& = (E”(x)+ + E”(x)−)/2 the ratio of Polε–synthesis on both strands. Plotted data was, when necessary smoothed by using a moving average of 3: the data point for each bin is an average of 7 points, the point at origin and the three points either side. Computational analysis was performed using the Apollo cluster computer at University of Sussex.
Identification of replication origins
Custom R scripts (available on request) were used to identify origins: polymerase usage ratio data from each strand (calculated without the assumption that 50% of the genome is replicated by Polε and 50% by Polδ) were smoothed by using a moving average of 3: the data point for each bin is an average of 7 points, the point at origin and the three points either side. The difference of each neighbouring data point was plotted against chromosome position. This dataset was further smoothed by applying a moving average of 3. The maximum of each positive peak was identified and peaks with a maximum below the lower 30th quartile of the dataset were ignored. Neighbouring peaks within 1200 bp (4 bins) were merged. The difference between the maxima and minima from the corresponding polymerase usage data (proportional with the areas under the peaks) was calculated as a measure of origin efficiency. Only origins that were present in both datasets (within plus minus 900 bps (3 bins) were considered and their efficiencies were averaged to generate a single origin efficiency, Efori.
Mapping origins by BrdU ChIP–Seq
YD18 cells were grown to exponential phase (0.2 ×106 /ml) at 25°C and synchronised at G2 phase by incubating these cell at 36°C for 3.5 hr cells. After adding bromo–deoxyuridine (0.5 μM) and hydoxyurea (10 mM) cell are further incubated at 25°C for 90 min, 1×108 cells were pelleted by the centrifugation and subjected to genomic DNA extraction. Subsequently BrdU–IP was performed as described in Xu et al, (2012)25.
Replication timing by marker frequency analysis
Cells (strain 501) were synchronised in G2 by elutriation (considered the least physiologically stressful method of synchronisation for fission yeast) concentrated into a volume of 200 ml and grown in fresh media at 27°C. Samples were taken at 5 minute intervals through S phase and analysed for DNA content by flow cytometry; mitotic index and septation by staining with DAPI and Calcoflor36. The population–averaged fraction of the genome replicated at each time–point was calculated from flow cytometry and septation index data. During flow cytometry sample preparation post–S phase S. pombe cells can separate. Consequently, during early time points after elutriation the 2N peak (in flow cytometry data) is predominantly pre–S phase cells, but in later time points the 2N peak starts to include post–S phase cells. We determined the proportion of pre–S phase (G2 and M), S phase and post–S phase (septum pinched in or two cells together) cells from the septation data. Flow cytometry was used to quantify the fraction of cells in the 2N peak and with a DNA content greater than 2N. Then, the septation index data was used to determine the proportion of the 2N peak that represented post–S phase cells. Briefly, if the proportion of G2 and M phase cells (septation data) was less than the proportion of cells in the 2N peak this difference could be attributed to either very early S phase or post–S phase cells. The post–S phase septation data allowed us to distinguish between these alternatives. In early time–points (20–85 min) the small proportion of post–S phase cells (≤7 %) were assumed to contribute to the 2N peak. In later time–points (90–120 min) the small proportion of G2 and M phase cells (≤10 %) were used to infer that the remaining cells in the 2N peak were post–S phase. Once the proportion of pre– and post–S phase cells in the 2N peak has been estimated, the flow cytometry data was used to determine the population–averaged fraction of the genome replicated at each time point. The DNA content signal from the 2N peak was assumed to correspond to a haploid genome content (copy number 1) and the signal from the 4N peak to a diploid genome content (copy number 2). This permitted calculation of the relative population–averaged genome copy number throughout the time–course. The reference sample was taken pre–replication, 45 mins after elutriation.
DNA was prepared from the elutriated reference sample and samples from within S phase, libraries were prepared and subjected to high–throughput sequencing as previously described27. The relative representation of each locus in the S phase samples was normalised to the percentage of total replication and to the unreplicated reference sample to provide an average percentage replication for each locus for each time point. To provide an unbiased replication timing map, S phase cell from an unperturbed exponentially growing culture were collected by FACS following fixation with 70% ethanol and subjected to marker frequency analysis using the sort–seq protocol previously described27).
Calculation of relative and absolute replication timing
The time course data was used to calculate a median absolute replication time (Trep) for each genomic locus as described previously27. Briefly, a sigmoidal function was fitted to the population–averaged fraction of the genome replicated at each time point for each genomic locus and Trep was determined as a time when the population–averaged fraction of the genome replicated was equal to 0.5. Times are shown relative to the approximate start of S phase, 50 minutes post–elutriation. Relative replication times and the distribution of replication termination sites were calculated from the Pu–seq fork direction data using custom scripts described previously5,28. Briefly, relative replication time was calculated from the integral of the percentage of leftward moving forks, assuming a constant average fork velocity across the genome. Termination frequency was calculated by using a finite difference approximation for estimating the derivative of the percentage of leftward moving forks.
Supplementary Material
1
2
3
Acknowledgments
We thank Jo Murray and Saed Mohebi for assistance with elutriation and Tom Kunkel for advice on polymerase mutation. AMC acknowledges Medical Research Council grant G1100074 and European Research Council grant 268788–SMI–DDR. CAN acknowledges Biotechnology and Biological Sciences Research Council grant BB/K007211/1.YD acknowledges a postdoctoral fellowship for research abroad from the Japan Society for the Promotion of Science.
Footnotes
Accession codes
The full list of origin locations, their calculated efficiencies and their relationship to previously identified origins is given in supplementary Table1. Names of supplementary files to visualise the full genome data for each panel in the figures is given in supplementary Table2. Formats are compatible with the IGV program available from the Broad Institute website. The data files are available at NCBI Gene Expression Omnibus under accession number GSE62108: (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=clmzquuwnrmdbkr&acc=GSE62108).
Competing interests statement
The authors declare that they have no competing interests in relation to this work.
References
- 1.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 3.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–84. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Hawkins M, et al. High-resolution replication profiles define the stochastic nature of genome replication initiation and termination. Cell reports. 2013;5:1132–41. doi: 10.1016/j.celrep.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryba T, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon AC, et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014;510:293–7. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta S, van Deursen F, de Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013;23:543–52. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Handa T, Kanke M, Takahashi TS, Nakagawa T, Masukata H. DNA polymerization-independent functions of DNA polymerase epsilon in assembly and progression of the replisome in fission yeast. Mol. Biol. Cell. 2012;23:3240–53. doi: 10.1091/mbc.E12-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24:602–12. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4949–54. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–6. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–4. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell. 2013;49:1010–5. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 22.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair. 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013;50:437–43. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, et al. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol. 2012;13:R27. doi: 10.1186/gb-2012-13-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiozaki K, Shiozaki M, Russell P. Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol. Biol. Cell. 1998;9:1339–49. doi: 10.1091/mbc.9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller CA, et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 2014;42:e3. doi: 10.1093/nar/gkt878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Retkute R, Nieduszynski CA, de Moura A. Mathematical modeling of genome replication. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;86:031916. doi: 10.1103/PhysRevE.86.031916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siow CC, Nieduszynska SR, Muller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:D682–6. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol. Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulrich HD, Takahashi T. Readers of PCNA modifications. Chromosoma. 2013;122:259–74. doi: 10.1007/s00412-013-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 1999;274:22283–8. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 34.Feng W, D’Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 2001;21:4495–504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 2014;21:664–70. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online Methods
- 36.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 37.Watson AT, Garcia V, Bone N, Carr AM, Armstrong J. Gene tagging and gene replacement using recombinase-mediated cassette exchange in Schizosaccharomyces pombe. Gene. 2008;407:63–74. doi: 10.1016/j.gene.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 38.Lipkin D, Talbert PT, Cohn M. The Mechanism of the Alkaline Hydrolysis of Ribonucleic Acids. J. Am. Chem. Soc. 1954;76:2871–2872. [Google Scholar]
- 39.Zhang Z, Theurkauf WE, Weng Z, Zamore PD. Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence. 2012;3:9. doi: 10.1186/1758-907X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–83. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3