High-throughput phenotyping of lung cancer somatic mutations (original) (raw)
. Author manuscript; available in PMC: 2017 Aug 8.
SUMMARY
Recent genome sequencing efforts have identified millions of somatic mutations in cancer. However, the functional impact of most variants is poorly understood. Here we characterize 194 somatic mutations identified in primary lung adenocarcinomas. We present an expression-based variant impact phenotyping (eVIP) method that uses gene expression changes to distinguish impactful from neutral somatic mutations. eVIP identified 69% of mutations analyzed as impactful and 31% as functionally neutral. A subset of the impactful mutations induces xenograft tumor formation in mice and/or confers resistance to cellular EGFR inhibition. Among these impactful variants are rare somatic, clinically actionable variants including EGFR S645C, ARAF S214C and S214F, ERBB2 S418T, and multiple BRAF variants, demonstrating that rare mutations can be functionally important in cancer.
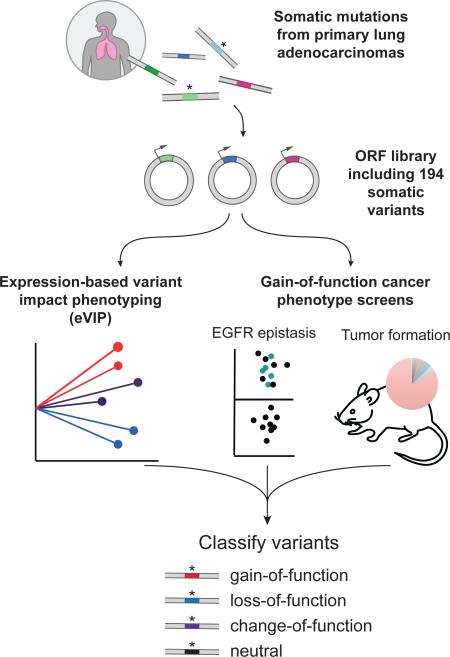
Graphical Abstract
INTRODUCTION
Cancer genome analysis is increasingly a part of clinical care (Frampton et al., 2013; Roychowdhury et al., 2011; Van Allen et al., 2014; Wagle et al., 2012), particularly in the setting of lung adenocarcinoma, in which somatic mutations in EGFR or translocations involving ALK or ROS1 confer tumor sensitivity to particular tyrosine kinase inhibitors (Kwak et al., 2010; Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004; Shaw et al., 2014). However, cancer-associated variants include not only “driver” mutations that are causally related to tumorigenesis but also “passenger” mutations that are inconsequential to tumor formation (Stratton et al., 2009). Because lung adenocarcinomas have high mutational burdens (Lawrence et al., 2013) and mutations can accumulate in non-expressed genes (Lawrence et al., 2013; Polak et al., 2015), it is likely that lung adenocarcinomas contain a high load of passenger mutations, including in genes of known importance in cancer.
Genome sequencing frequently identifies variants of unknown significance (VUS), even in disease-associated genes (Kilpivaara and Aaltonen, 2013; Taylor et al., 2015). For example, approximately one third of TCGA lung adenocarcinoma samples with non-synonymous mutations in EGFR harbor mutations at non-recurrently mutated sites. This proportion rises to 45% of observed EGFR mutations across cancers (www.tumorportal.org). Information on whether these VUS have a similar impact as previously characterized mutations will be critical to guide clinical decision-making for patients whose lung tumors harbor these mutations. Given that the number of genes associated with cancer is continuing to increase with increasing power of genomics studies (Lawrence et al., 2014), development of technologies to assess the functional consequence of individual mutations at scale is warranted.
Recent advances in genetic perturbation tools have allowed assessment of wild-type gene function at scale, yielding important insights into tumor biology (Boehm et al., 2007; Ebert et al., 2008), drug resistance (Johannessen et al., 2013; Shalem et al., 2014; Wilson et al., 2015), and signal transduction (Berns et al., 2004; Parnas et al., 2015). Furthermore, in vivo experiments enable the discovery of oncogenes and tumor suppressor genes in regions of chromosomal imbalance and have been successfully utilized to discover important genes in hepatocellular carcinoma (Sawey et al., 2011; Zender et al., 2008), lymphoma (Bric et al., 2009), and ovarian cancer (Dunn et al., 2014). Single-gene studies have provided valuable insight into somatic genetic variants in genes such as FLT3 (Frohling et al., 2007), and new technologies will increasingly allow such single-gene studies to approach saturation (Emery et al., 2009; Kitzman et al., 2015; Melnikov et al., 2014). Finally, large-scale site-directed mutagenesis has recently been employed to create the Human Mutation ORFeome 1.1, a library of thousands of germline variants, which was recently leveraged to understand the consequences of genetic variation on protein-protein interactions (Sahni et al., 2015).
Given these advances, we hypothesized that it might now be possible to simultaneously assess the molecular and phenotypic consequences of hundreds of somatic mutations across genes of diverse function.
RESULTS
A mutated ORF library of lung adenocarcinoma
We prioritized 53 genes for study based on mutation frequency, relevance to known lung adenocarcinoma pathways and availability of open-reading frame (ORF) plasmid templates (Table S1). Exome sequencing of 412 primary lung adenocarcinomas identified 518 unique missense mutations or in-frame insertions or deletions in these 53 genes (TCGA, 2014) (Imielinski et al., 2012). We used site-directed mutagenesis to generate mutant clones and transferred ORFs to lentiviral expression vectors. Overall, we successfully generated 194 mutant expression constructs. Adding wild-type controls and other genes used as expression controls or pathway references, we utilized 352 different lentiviral expression constructs (Table S1).
We first used the SIFT (Ng and Henikoff, 2003), PolyPhen2 (Adzhubei et al., 2010), and MutationAssessor (Reva et al., 2011) computational algorithms to predict which somatic mutations might impact protein function. While these methods agreed for 66/103 predictions (64%), they disagreed on multiple variants including those in clinically relevant genes such as EGFR and KRAS (Figure S1A). To resolve such discrepancies and corroborate these in silico predictions, we sought to experimentally determine the functional impact of the selected mutations.
Development of an expression-based variant impact phenotyping assay
Typically, measuring the impact of mutations in a given gene requires a distinct assay or measurement of a biomarker specific for each gene under study. However, previous reports demonstrate the feasibility of using gene expression signatures as “fingerprints” of molecular function (Lamb et al., 2006). Thus, we reasoned that comparing gene expression changes induced by wild-type and mutated alleles of the same gene could provide functional insight without requiring prior knowledge of gene function. While we expect that different cellular contexts could in theory yield different signatures for certain variants, we elected to start in a specific context, A549 lung adenocarcinoma cells, as an initial proof of concept.
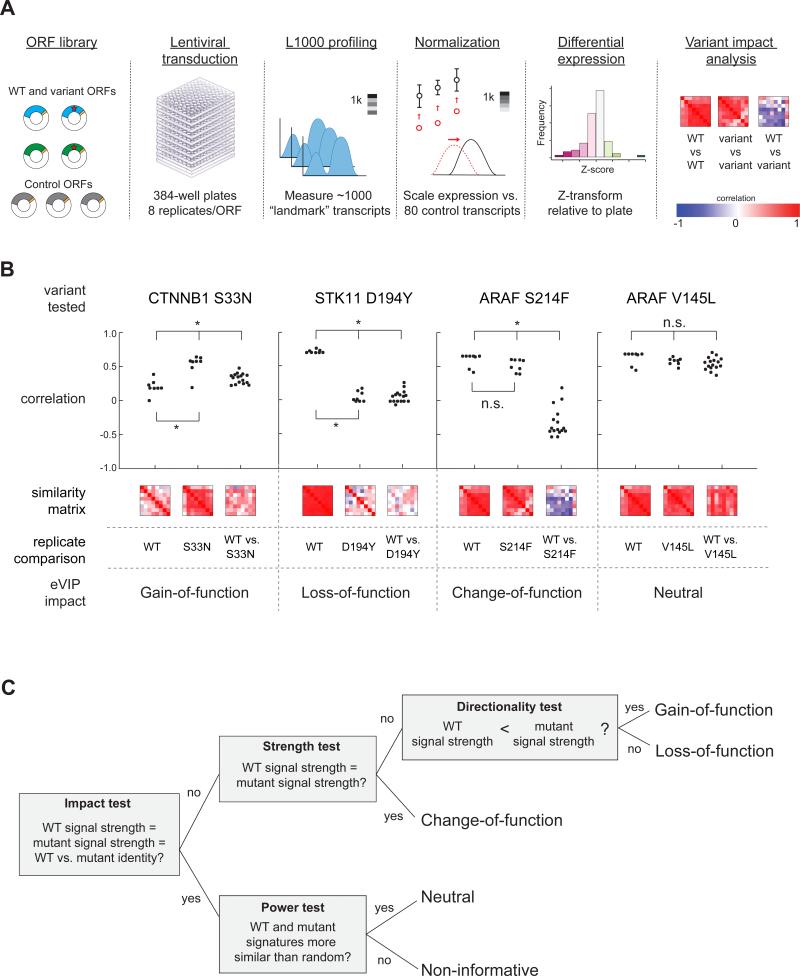
We introduced the ORF library into A549 cells via lentiviral transduction in arrayed format (1 ORF/well) with 8 biological replicates per ORF (Figure 1A). 96 hours later, gene expression of 978 transcripts was assayed using L1000 profiling (Peck et al., 2006). Each L1000 profile was normalized to a set of housekeeping transcripts (Figure 1A). Differential gene expression signatures were then generated by transforming each transcript level to a robust Z-score relative to the median background expression in the parental cell line (Figure 1A). The complete Z-scored data is supplied in File S1.
Figure 1. Expression-Based Variant Impact Phenotyping.
(A) Overview of the experimental pipeline from reagent generation to eVIP analysis.
(B) Dot-plot and heat map representation of replicate consistency (WT vs. WT or variant vs. variant) comparisons and signature identity (WT vs. variant) comparisons. Correlation is measured by a weighted connectivity score (wtcs). *, adjusted p < 0.05. n.s., adjusted p > 0.05.
(C) Schematic of the decision tree-based eVIP algorithm. The first test (“impact test”) outputs a single Bonferroni-adjusted p value indicating likelihood of mutation impact. For impactful mutations, the next tests are used to determine the directional impact of the mutations. For mutations found to be non-impactful, a “power test” assesses whether the two signatures are similar to one another due to real signal, or due to noise.
See also Figure S1, Table S1, and File S1.
Next, we compared each mutant signature to its respective wild-type control signature to test the null hypothesis that the two signatures are identical. We measured both the strength of signal and the signature identity (which transcripts are altered). Replicate consistency is a quantitative measure of signal strength with a greater dynamic range than direct calculation of signal strength from Z scores alone (Figure S1B). ORFs that induce weak gene expression changes have inconsistent differential expression changes and thus have poor self-correlation across replicates. In contrast, ORFs that induce strong gene expression changes have consistent changes with large magnitude and high correlation across replicates.
Some mutated ORFs, such as CTNNB1 S33N, induced signatures with an increase in replicate consistency relative to the cognate wild-type ORF signature (Figure 1B). We concluded that this directionality (mutant>WT) indicates a gain-of-function (GOF) mutation. In contrast, other mutated ORFs, such as STK11 D194Y, induced signatures with decreased replicate consistency compared to wild-type (Figure 1B), suggesting loss-of-function (LOF) impact on gene function.
In cases in which the replicate consistency of the wild-type and mutant signatures did not differ, as for ARAF S214F and V145L relative to wild-type ARAF(Figure 1B), we compared signature identity by calculating the correlation between the differentially expressed transcripts in the wild-type signature and those in the mutant signature. ARAF S214F induced substantially different gene expression changes than wild-type ARAF whereas gene expression induced by ARAF V145L was indistinguishable from that induced by wild-type ARAF (Figure 1B). Thus, S214F represents an impactful variant of ARAF whereas V145L represents a neutral variant in this context. We classified mutants with similar replicate consistency to wild-type but with significantly different signature identity as change-of-function (COF).
We combined the replicate consistency and signature identity analyses into a decision tree algorithm and built a computational pipeline for expression-based Variant Impact Phenotyping, or eVIP, to assess the impact and genetic directionality of each mutation (Figure 1C, Figure S1C).
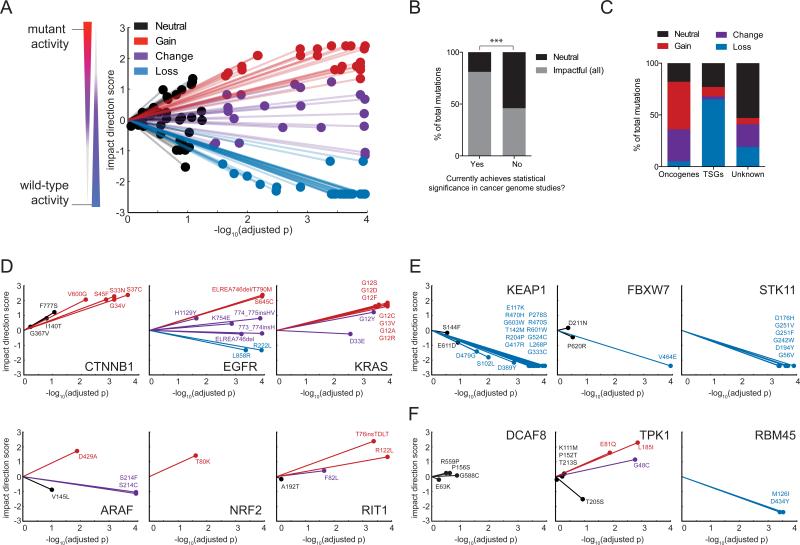
Starting with 194 mutant alleles, we considered the 110 highest quality comparisons in which both wild-type and mutant signatures were available, had high infection efficiency, and in which all replicates passed L1000 quality control. Of the 110 mutant alleles, 75 (69%) mutations were found to significantly impact wild-type function under these experimental conditions and after multiple-hypothesis correction (Benjamini-Hochberg false discovery rate (FDR) < 5%) (Figure 2A and Table S2). It is likely that this value significantly overestimates the true fraction of impactful mutations in lung cancer due to the non-random selection of genes and mutations in this study.
Figure 2. eVIP Classifications for 110 Lung Cancer Somatic Mutations.
(A) A “sparkler” plot representation of eVIP predictions. Each variant allele with an eVIP prediction is represented as a point. The x-axis indicates the –log10 (adjusted p) of a Kruskal-Wallis test comparing wild-type and mutant ORF replicate consistency and signature identity. The y-axis is the “impact direction score,” the absolute value of which is equal to the –log10 (adjusted p) of a Wilcoxon test directly comparing wild-type and mutant ORF replicate consistency. The sign of the impact direction score is positive if the mutant ORF replicate consistency is greater than the wild-type replicate consistency and negative if the mutant ORF replicate consistency is less than the wild-type ORF replicate consistency. The line connecting each point and the graph origin is drawn to emphasize that longer distance from the origin implies more confidence in the prediction.
(B) Enrichment of impactful variants in genes found to be significantly mutated cancer genome studies. GOF, COF and LOF predictions are all considered impactful. ***, p < 0.0001 by Fisher's Exact test.
(C) Distribution of eVIP calls in known oncogenes, known tumor suppressor genes (TSGs), or genes of unknown function.
(D-F) Gene-specific sparkler plots for known oncogenes (D), known tumor suppressor genes (E), and genes of unknown function (F). The tested variant allele is labeled and colored based on the eVIP prediction. Coloring is as in (A).
See also Table S2, Table S3, Table S4, and File S2.
To determine to what extent these predictions are correct, we took multiple approaches. First, to estimate the false positive rate of eVIP, we performed mock comparisons from an independent 24-replicate eVIP experiment (File S2). We sampled 8 replicates from one ORF as a “mock WT” signature and compared to 8 independent replicates from the same ORF as a “mock mutant” signature. Because all replicates actually represent the same ORF, any significant differences between these comparisons represent false positives. Performing 1000 iterations of this analysis, 4.74% of comparisons were falsely called impactful, suggesting the eVIP FDR cutoff of 5% is well-calibrated and reflects the empirical false-positive rate.
Second, we assessed the sensitivity of eVIP by comparing the eVIP predictions to previous data for 21 mutations for which prior experimental evidence was available. eVIP correctly inferred mutation impact for 100% of 21 benchmark alleles, suggesting eVIP is highly sensitive (Table S3). In 19 of these benchmark cases, eVIP returned the correct directionality of the mutation (GOF/COF vs. LOF). In limited cases, eVIP may have been underpowered to resolve true positives. For instance, we note that the EGFR L858R variant was correctly determined to be impactful but incorrectly called a LOF mutation. To investigate this discrepancy, we analyzed data of greater statistical power (24-replicate experiment vs. the 8-replicate main experiment) and found that this higher-powered experiment resolved EGFR L858R as a COF mutation (Table S4).
Third, we asked whether the frequency and predicted genetic directionality of impactful mutations was related to the categories of the genes under study. 81% (57/70) of mutations in genes that were considered significantly mutated in recent cancer genome studies (Imielinski et al., 2012; Lawrence et al., 2014; TCGA, 2014) were impactful, whereas only 46% (18/39) of mutations in genes with lower mutational frequencies were impactful (Figure 2B). As expected, oncogenes were enriched for GOF and COF mutations and tumor suppressor genes were enriched for LOF mutations (Figure 2C). These patterns are readily evident when visualizing the eVIP data on gene-specific “sparkler” plots (Figure 2D-F). We note that, surprisingly, this method was able to detect impactful mutations in oncogenes, including KRAS, despite the presence of an endogenous, activating KRAS mutation in A549 cells. Unlike oncogenes (Figure 2D), known tumor suppressors were enriched for LOF mutations (Figure 2E). Patterns of mutation impact in genes of unknown significance in cancer could be similarly stratified by enrichment for either GOF or LOF mutations (Figure 2F).
To further evaluate the eVIP approach, we compared functional impact predictions with predictions from the computational methods SIFT, Polyphen2 HumDiv, and MutationAssessor (Adzhubei et al., 2010; Ng and Henikoff, 2003; Reva et al., 2011). For 66 variants that all three computational methods agreed on (52 impactful, 14 neutral), eVIP also agreed on the functional impact of 56 variants. Only eVIP and PolyPhen2 HumDiv correctly predicted the impact of 21 literature benchmarks. However, PolyPhen2 HumDiv may not have good specificity given that variants were predicted to be impactful 81% of the time, in contrast to 69% predicted to be impactful by eVIP, suggesting eVIP may have good specificity while also having high sensitivity.
Widespread loss-of-function missense variants in tumor suppressor genes
A major challenge in the interpretation of lung cancer genomes involves discriminating impactful from neutral missense mutations in tumor suppressor genes (TSG), since such genes display a widely distributed pattern of mutation throughout the gene (Davoli et al., 2013). For the missense mutations studied in seven known or putative tumor suppressor genes (DOK1, FBXW7, KEAP1, MAX, PPP2R1A, RB1, STK11), 77% of mutations were predicted to be impactful. However, the distribution of impactful mutations varied by gene; 92% of mutations in KEAP1 and STK11 (23 of 25) impacted gene function compared to wild-type (Figure 2E).
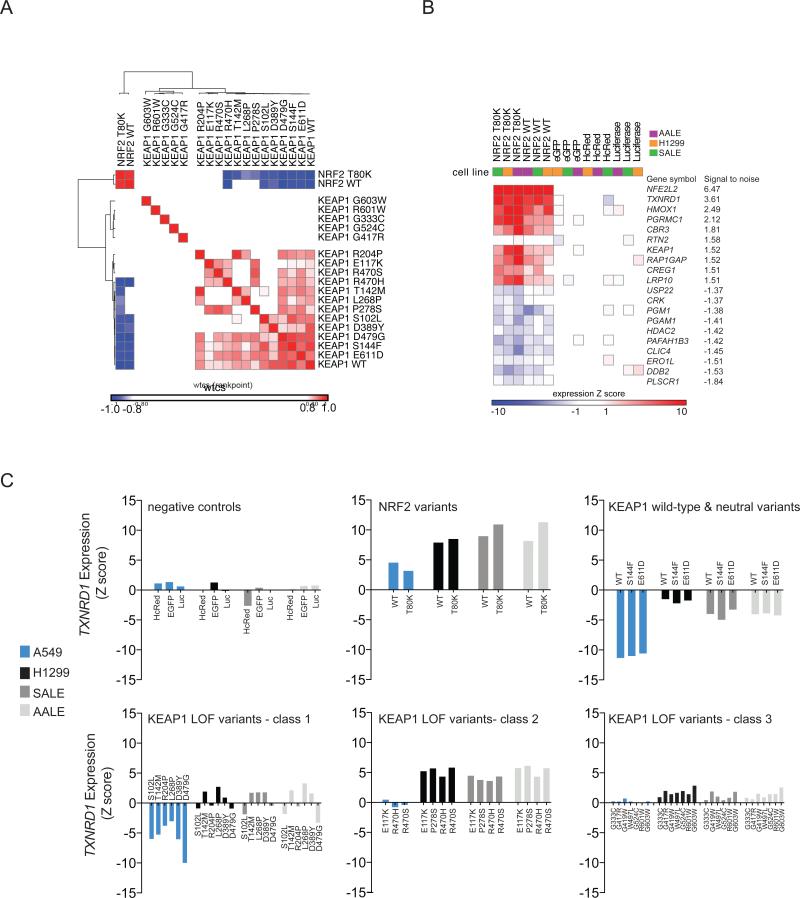
KEAP1 encodes an E3 ubiquitin ligase that negatively regulates the protein level of the NRF2 transcription factor encoded by NFE2L2 (Jaramillo and Zhang, 2013). Because A549 cells express elevated levels of NRF2 as a consequence of a KEAP1 G333C mutation, we expected that expression of a wild-type KEAP1 ORF should suppress NRF2 levels in this cell line. As expected, the transcriptional signature of wild-type KEAP1 and two KEAP1 variants predicted by eVIP to be neutral (E611D and S144F) were the most anticorrelated with NRF2 overexpression (Figure 3A). In contrast, the expression signatures induced by KEAP1 variants G603W, R601W, G333C, G524C, and G417R showed no correlation, positive or negative, with the NRF2 expression signature, indicative of severe loss of KEAP1 function. All other KEAP1 variants determined to be impactful by eVIP still retained some ability to regulate NRF2 as judged by the mutant signatures’ correlation with the wild-type KEAP1 signature and anti-correlation with the NRF2 signature (Figure 3A). These mutations are likely to represent hypomorphic variants of KEAP1 and overlap with reported hypomorphs of KEAP1 recently characterized by Hast and colleagues (Hast et al., 2014).
Figure 3. Hypomorphic and Dominant-negative KEAP1 Variants Identified by Gene Expression Profiling.
(A) Hierarchical clustering of KEAP1 expression signatures in A549 cells. The similarity matrix was computed using the weighted connectivity score (wtcs) as the similarity metric.
(B) Two-class comparison of NRF2 ORF signatures versus control (EGFP, HcRed, Luciferase) signatures across three KEAP1 wild-type cell lines (AALE, SALE, H1299). The top transcripts up- or down-regulated by NFE2L2 expression were determined by a signal-to-noise statistic.
(C) Expression of the direct NRF2 transcriptional target TXNRD1 is shown as a biosensor of NRF2 and KEAP1 activity. Upper panels show NRF2 variants and KEAP1 wild-type and neutral variants. The lower panels show three classes of loss-of-function KEAP1 variants identified by hierarchical clustering of expression profiles generated in four cellular contexts: A549 (KEAP1 mutant) and three KEAP1 wild-type cell lines, H1299, SALE, and AALE.
See also Figure S2 and File S3.
We next assessed whether different cellular contexts would provide further resolution of these mutation assessments. We introduced the entire KEAP1 variant series into three KEAP1 wild-type cellular contexts (H1299 lung cancer cells, and AALE and SALE immortalized, non-transformed lung epithelial cells) (File S3), to determine whether dominant-negative mutations could be resolved. The top upregulated genes upon NRF2 overexpression in all KEAP1 wild-type cell lines were the known NRF2 transcriptional targets TXNRD1 and HMOX1 (Malhotra et al., 2010) (Figure 3B). Using these two transcripts as biosensors of NRF2 activity, KEAP1 expression signatures were clustered (Figure S2). This analysis revealed a cluster of alleles with KEAP1 wild-type activity, plus three different classes of impactful KEAP1 variants. The first class consisted of KEAP1 hypomorphic variants that retained the ability, to some extent, to regulate NRF2 in A549 (Figure 3C). The second class consisted of 4 KEAP1 variants (R470S, R470H, P278S, and E117K) that displayed a strong dominant-negative effect whereby expression of these alleles in all three KEAP1 wild-type contexts induced upregulation of NRF2 target genes, indicative of NRF2 stabilization (Figure 3C). The third class, severe loss-of-function KEAP1 variants, was also able to exert a dominant-negative activity in the KEAP1 wild-type cell lines (Figure 3C), although to a lesser degree than the class 2 mutations.
Despite the observed dominant-negative activity in vitro, analysis of the 19 tumors from which these KEAP1 variants were identified revealed LOH accompanying 17 of 19 KEAP1 mutations, including the four mutations showing dominant-negative activity in vitro, indicating that full genetic loss of KEAP1 provides the greatest selective advantage to human tumors, even for alleles with dominant-negative activity.
Identification of mutations epistatic to EGFR
A remarkable diversity of rare mutations in EGFR, KRAS and other oncogenes in lung cancer have been identified, and it remains unclear which, if any, function similarly to canonical hotspot mutations. eVIP predicted 91% of the tested variants in ARAF, EGFR, KRAS, NRAS, and RIT1 to be impactful, but one limitation of eVIP is that it does not directly assess which of these variants are oncogenic. To further characterize the phenotypic impact of rare oncogenic mutations, we utilized complementary in vitro and in vivo phenotypic assays to provide additional evidence that specific variants activate the EGFR/RAS pathway.
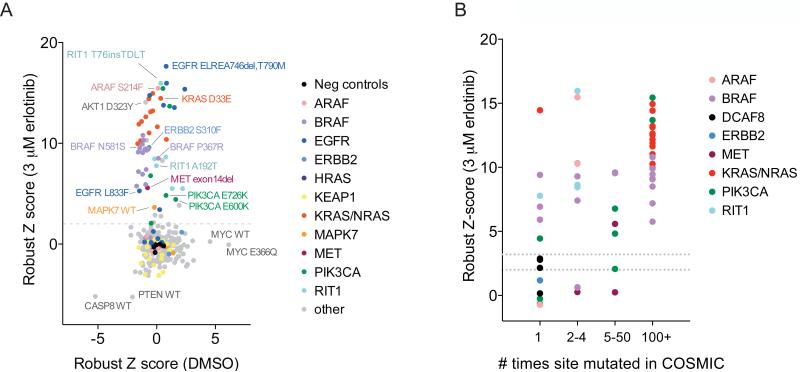
First, we leveraged a recently developed erlotinib-rescue assay in PC9 lung adenocarcinoma cells (Sharifnia et al., 2014) to determine which mutations represent gain-of-function mutations that are epistatic to EGFR. PC9 cells harbor an activating EGFR exon 19 in-frame deletion and are naturally sensitive to the EGFR inhibitor erlotinib. Expression of variants such as mutant KRAS that re-activate downstream signaling pathways can rescue the erlotinib-induced lethality in this cell type (Sharifnia et al., 2014).
We determined the ability of each ORF to rescue erlotinib sensitivity at two erlotinib doses after 72 hours of treatment. Cell viability across replicate conditions and in both erlotinib doses was highly correlated (Figure S3A-S3E). 62 of 351 ORFs (17.7%), representing variants of 14 proteins, rescued cell viability in 3 μM erlotinib, including numerous mutant variants of KRAS, EGFR, RIT1, and BRAF (Z score > 2; Figure 4A). We retested 42 ORFs that had scored in at least one erlotinib dose across a wider range of 5 erlotinib concentrations. 27/27 hits with primary screen Z scores greater than 3 and 9/15 hits with Z scores between 2 and 3 were confirmed upon retesting (Table S5 and Figure S3F).
Figure 4. Orthogonal Assays in Different Cellular Contexts Identify Oncogenic EGFR Pathway Mutations.
(A) Cell viability of PC9 cells after mutant allele library infection and 72 hours of treatment with 3 μM erlotinib or DMSO. Data shown are the average robust Z scores of two replicates per ORF condition. A dashed line indicates the threshold used to select ORFs for validation (Z > 2).
(B) Relationship between mutation frequency in COSMIC (x-axis) and ability to rescue cell viability in erlotinib (y-axis). Two dashed lines indicate the Z score thresholds used to select ORFs for validation (Z > 2) and the threshold at which all ORFs retested in validation (Z > 3).
See also Figure S3 and Table S5.
As expected, known resistance variants of EGFR, L858R/T790M and 746delELREA/T790M, conferred erlotinib resistance whereas known sensitive alleles L858R and exon 19 deletions, did not. EGFR exon 20 insertion alleles 773_774insH, 769_770insASV, and 774_775insHV conferred resistance to erlotinib (Figure S3E, S3F), in agreement with previous findings (Greulich et al., 2005; Oxnard et al., 2013). Mutations frequently observed in the COSMIC database were more likely to score than rare mutations (Figure 4B); 22 of 22 mutations with a frequency in COSMIC greater than 100 scored in the assay versus 6 of 14 mutations observed only once in COSMIC (p < 0.0001 by Fisher’s exact test). However, rare “n of 1” mutations also scored in this assay, demonstrating that rare mutations can be functionally significant.
Identification of tumor-promoting mutations by multiplexed in vivo screening
To directly test which alleles promote tumor formation in vivo, we developed a multiplexed xenograft tumorigenesis assay of immortalized human lung epithelial cells sensitized to read out variants in the EGFR/RAS pathway. Our preliminary work found that immortalized small-airway epithelial cells harboring an activating YAP1 variant (SALE-Y cells) are incapable of forming tumors larger than 0.2 cm3 in mice up to at least 120 days, but are rendered tumorigenic via the introduction of activating variants in EGFR, BRAF, or MAP2K1 (Figure S4A). The reciprocal analysis showed that stably transduced SALE-EGFRL858R cells failed to form tumors in combination with the control ORFs HcRed and Renilla, but robustly formed tumors after transduction with activated YAP1 (Figure S4A). Based on these data, we chose the SALE-Y cellular context for screening, with the goal of identifying activating mutations in the EGFR/RAS pathway.
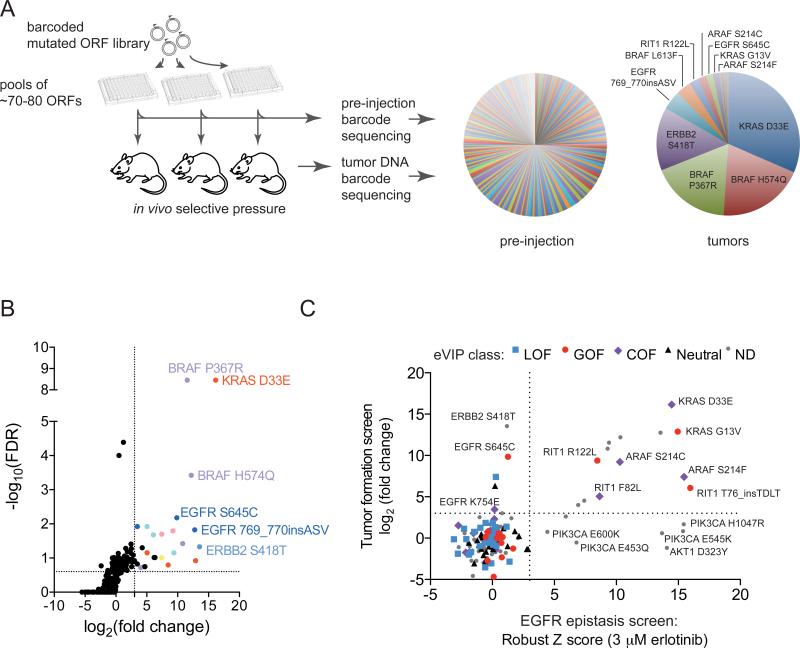
We introduced each of the barcoded alleles into SALE-Y cells in arrayed format, pooled in groups of ~70-80, and injected cells subcutaneously into immunocompromised mice (Figure 5A). We excluded most known oncogenic alleles from experimental pools so that these expected strong alleles would not dominate the tumor cell population. In addition, we performed the experiment using two different pool compositions; one composition contained pools including all test alleles and the other had the same 8 pools but omitted alleles that we reasoned were likely to be oncogenic. Overall, 92/96 injection sites formed tumors, with tumor latencies ranging from 11 days to 74 days.
Figure 5. A Multiplex In Vivo Tumor Formation Screen for Identification of Activated Oncogenes.
(A) Left, experimental schematic showing screening workflow. Right, pie charts showing the median corrected reads per million (RPM) per ORF in pre-injection and tumor samples for all pools in the tumor experiment. Because ORFs were assayed in different pools, the proportion of each ORF on the pie chart may not reflect the actual relative levels of oncogenic activity.
(B) One-sided volcano plot showing distribution of ORFs from all pools. Each datapoint represents data generated from all pre-injection and tumor replicates for a given ORF. The log2 fold-change (x-axis) was calculated by comparing the median corrected RPM of each ORF from the pre-injection samples to the median corrected RPM in tumor samples.
(C) Plot showing relationship between ORF variants across functional and expression-based screens. All alleles with both tumor formation and EGFR epitasis data were plotted; alleles not analyzed by eVIP are indicated by “ND” (not determined). The colored shapes indicate the predicted mutation impact as assessed by eVIP in A549 cells.
See also Figure S4, Table S6, and Table S7.
To determine which ORFs conferred tumor-forming capacity to SALE-Y cells, we harvested tumor DNA and PCR-amplified and sequenced ORF barcodes from each tumor. ORFs with poor pre-injection representation were excluded from further analysis (Figure S4B). By identifying barcodes that were significantly increased in abundance in tumors versus the pre-injection pools, the top hits were non-canonical alleles in known EGFR/RAS pathway genes such as BRAF H574Q, BRAF P367R, KRAS D33E, EGFR S645C, ERBB2 S418T, and RIT1 R122L (Figures 5A, 5B, and Table S6).
The hits identified in both the EGFR epistasis screen and tumor formation screens showed significant overlap (p < 0.0001 by Fisher's exact test; Figure 5C). A summary of the mutational impact of each variant as assessed by eVIP, the EGFR epistasis screen, and tumor formation screens is shown in Table S7. All mutations that scored in both the EGFR epistasis screen and tumor formation screen and were characterized by eVIP were classified as gain-of-function or change-of-function mutations by eVIP (ARAF S214C, ARAF S214F, KRAS D33E, KRAS G13V, RIT1 F82L, RIT1 R122L, and RIT1 T76insTDLT). No neutral or loss-of-function mutations scored in both of the functional screens. 33/34 (97%) and 29/30 (97%) of mutations called neutral or loss-of-function by eVIP, respectively, failed to score in either of the two functional screens (Figure 5C). Seven mutations scored in EGFR epistasis but did not promote tumor growth; five of these were PIK3CA or AKT1 variants. Five mutations induced tumor formation but did not score in the EGFR epistasis screen; three of these corresponded to EGFR or ERBB2 variants that may have been inhibited themselves by erlotinib.
Integrative functional and expression analysis of somatic variants
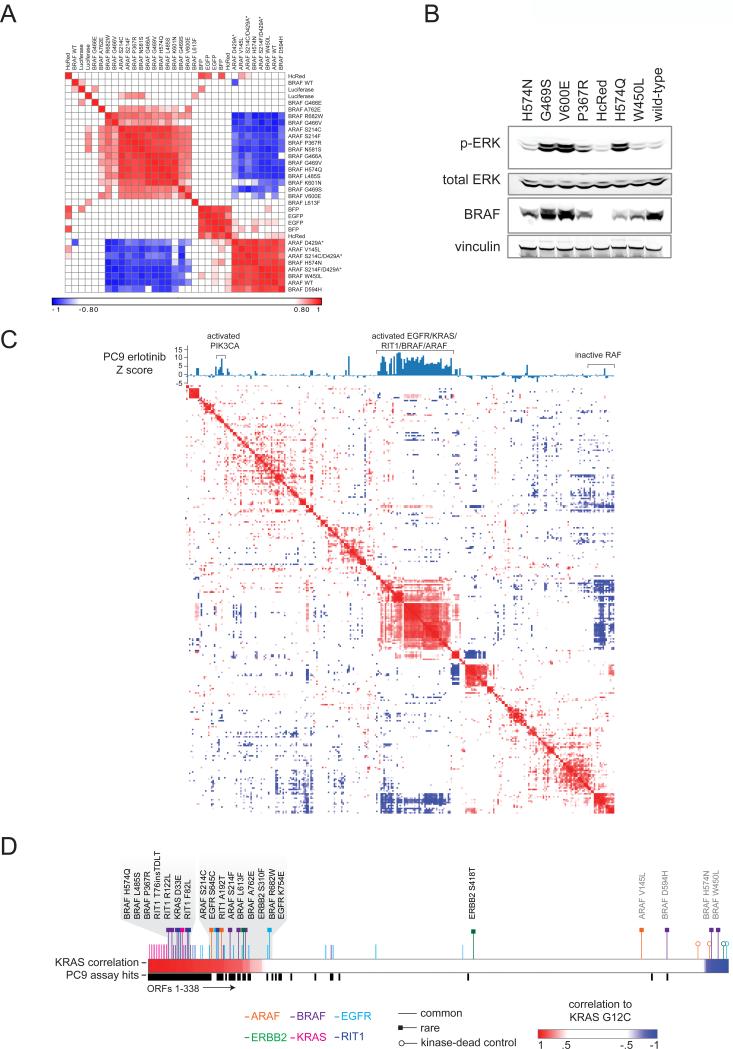
ARAF S214F and S214C were originally identified in a RAF inhibitor exceptional responder study (Imielinski et al., 2014). The expression signatures induced by these ARAF mutants are highly correlated to those induced by canonical BRAF mutants (Figure 6A). Both ARAF S214C and S214F robustly rescued cell viability in the presence of erlotinib and induced tumor formation. These activities were clearly kinase-dependent as each mutation in cis with a kinase-inactivating D429A mutation failed to induce tumor formation or erlotinib-resistance. Notably, ARAF V145L failed to score in any of the three assays, suggesting it is a passenger mutation.
Figure 6. Integration of Expression Signatures and Functional Data Identifies Rare Activating Mutations in the RAS/MAPK pathway.
(A) Heat map showing hierarchical clustering of expression signatures in A549 cells. Similarity of signatures was compared using the weighted connectivity score (wtcs) and transformed to a rankpoint distribution with the best correlation set to 1 and best anti-correlation set to −1. Asterisks indicate engineered kinase-dead ARAF variants (not found in human cancer).
(B) Western blot of A549 cells expressing wild-type BRAF or BRAF variants, or control vector (HcRed). The primary antibodies used are indicated on the left. p-ERK, phosphorylated Thr202/Thr204 ERK1/2.
(C) Hierarchical clustering of a similarity matrix of all A549 expression signatures in the study. Similarity of signatures was computed as in (A). A bar chart above the heatmap indicates the average robust Z score achieved by each respective ORF in the PC9 EGFR epistasis screen.
(D) Expression signature analysis in A549 cells showing all ORFs ranked from left to right in order of correlation to the KRAS G12C signature. Canonical alleles of EGFR/RAS pathway oncogenes and rare alleles induce signatures highly similar to KRAS G12C (left, gray shading), with the exception of ARAF V145L, determined to be neutral by eVIP, and the three apparently inactive BRAF mutants described in the text (right side, gray text). An additional track shows allele performance in the EGFR epistasis assay (black bars).
The majority of BRAF-mutants clustered with the known BRAF activating variant, V600E (Figure 6A). However, three BRAF variant signatures (W450L, H574N, D594H) were highly similar to the kinase-dead ARAF signature, suggesting these BRAF variants lack catalytic and/or other activity. BRAF D594 mutants have been previously described as kinase-inactive but able to activate MAPK signaling in RAS mutant cells (Heidorn et al., 2010). However, because our experiment was performed in _KRAS_-mutant A549 cells, the expression data presented suggest other factors in addition to RAS mutation status may determine the functional output of kinase-dead BRAF. Western blot analysis of A549 cells expressing BRAF variants confirmed that H574N and W450L fail to induce ERK phosphorylation whereas other rare variants H574Q and P367R induce higher ERK phosphorylation levels than that induced by expression of wild-type BRAF (Figure 6B).
The majority of activating variants in EGFR, BRAF, and ARAF, as well as those in the small GTPases KRAS and RIT1 (Berger et al., 2014), induced expression signatures which clustered together and were distinct from the wild-type or inactive RAF allele signatures (Figure 6C and Figure 6D). The activated EGFR-pathway oncogene cluster correlated with the high scoring ORFs from the PC9 EGFR epistasis screen (Figure 6C and Figure 6D). The correlation across cellular contexts is encouraging and suggests that in the future, expression profiling alone may be sufficient to identify activating mutations in this pathway.
In addition to previously characterized EGFR variants such as L858R and exon 19 deletions, eVIP identified two rare non-canonical EGFR variants, S645C and K754E, as gain-of-function mutations. Of note, SIFT, PolyPhen2, and MutationAssessor were in disagreement on the functional impact of these EGFR variants. In agreement with the eVIP predictions, both EGFR S645C and EGFR K754E promoted tumor formation in vivo. However, unlike other EGFR variants that are sensitive to erlotinib, these two variants were less sensitive to erlotinib than wild-type EGFR (Figure S3F), suggesting that patients with tumors harboring these particular alleles might not benefit from treatment with this inhibitor.
MEK inhibition overcomes erlotinib resistance induced by EGFR-pathway oncogenic mutations
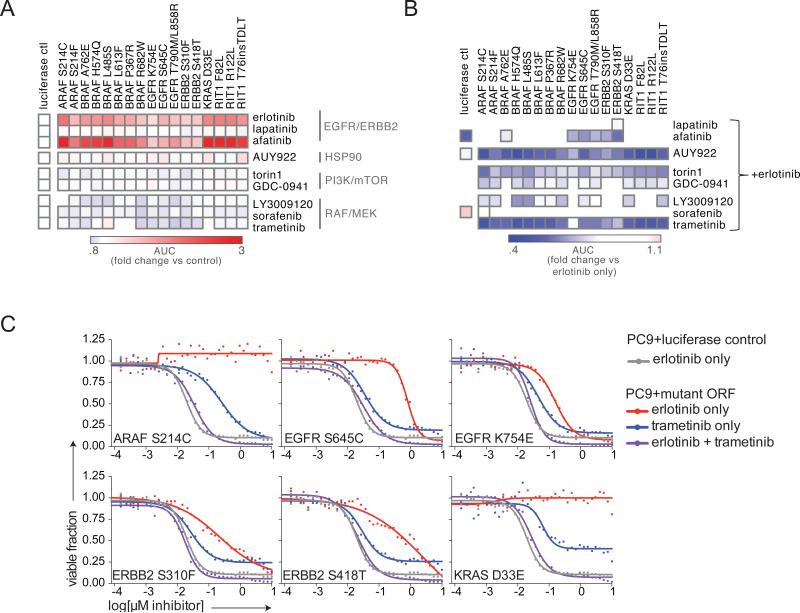
The rare or non-canonical variants of ARAF, BRAF, EGFR, ERBB2, KRAS, and RIT1 identified in the functional assays above induce expression signatures highly correlated with mutant KRAS activation (Figure 6D), suggesting these alleles activate RAS/MAPK signaling. We hypothesized that inhibition of downstream nodes in the EGFR/RAS pathway might overcome erlotinib resistance induced by these oncogenic alleles. To investigate the sensitivity of these alleles to targeted therapies, we generated 16 isogenic PC9 stable cell lines expressing the mutant alleles plus two control isogenic lines (Figure S5). We assayed the response of the cell lines to erlotinib or 8 different small molecule inhibitors targeting EGFR/ERBB2, HSP90, MEK, and PI3K/mTOR (Figure 7A).
Figure 7. Sensitivity of Rare Oncogenic Mutations to MEK Inhibition.
(A) Viability of PC9 stable isogenic cell lines after treatment with the small molecules shown for 96 hours. 36-point dose response curves were performed and curves plotted in GraphPad Prism. Data shown are the ratio of the area-under-the-curve (AUC) of the mutant PC9 line vs. the PC9-luciferase negative control.
(B) Viability of PC9 stable isogenic cell lines after treatment with the small molecules shown in combination with erlotinib. Inhibitors were delivered in a 1:1 molar ratio across a 36-point dose-response curve. Data shown are the ratio of the AUC of the mutant PC9 line in the combination treatment vs. the AUC of the mutant PC9 in erlotinib only. Note that EGFR K754E appears to be the least sensitive to trametinib inhibition but this is actually due to EGFR K754E conferring the least resistance to trametinib, as shown in panel (C).
(C) Dose-response curves of PC9 stable isogenic cell lines expressing EGFR, ERBB2, or KRAS variants. Data shown are the same curves used to generate the heat maps in panel (B).
See also Figure S5.
As expected from the primary EGFR epistasis screen, the stable isogenic cell lines exhibited resistance to erlotinib treatment (Figure 7A). All lines also exhibited cross-resistance to the EGFR inhibitor afatinib, but not to other inhibitors tested (Figure 7A). However, co-treatment with erlotinib and trametinib could overcome the erlotinib resistance in all lines, indicating that the activity of these rare oncogenic alleles is dependent on re-activation of MEK (Figure 7B, 7C). Co-treatment with erlotinib and the mTOR inhibitor torin1 could also largely overcome erlotinib resistance, indicating that all tested mutants act both upstream of MEK and also mTOR to promote survival in erlotinib (Figure 7B). The response of all lines to HSP90 inhibition by AUY922 was not altered by expression of any allele or by combination treatment with erlotinib.
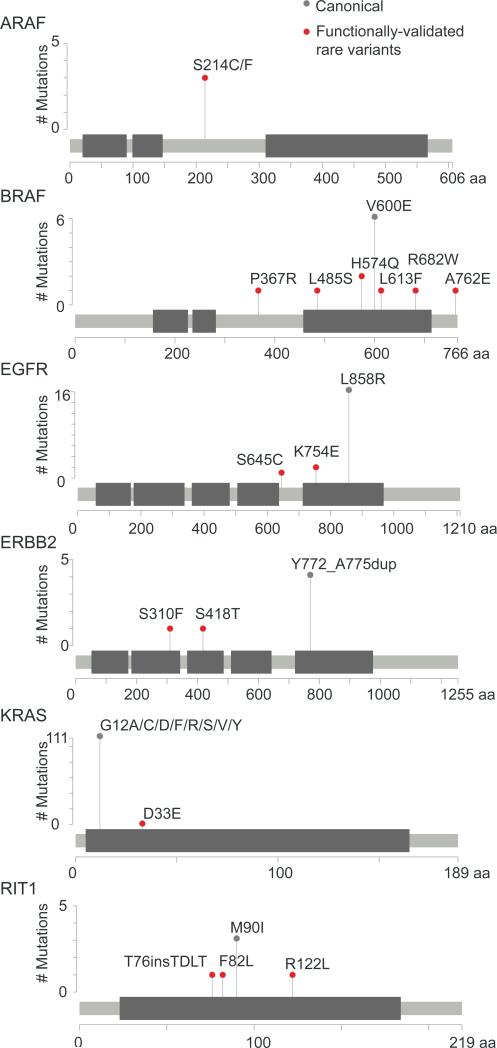
Taken together, we validated 16 rare oncogenic variants in ARAF, BRAF, EGFR, ERBB2, KRAS, and RIT1 as activating mutations that both induce tumor formation and confer resistance to erlotinib in a MEK-dependent manner (Figure 8), indicating that MEK inhibition should be explored as a potential therapeutic strategy for patients with tumors harboring these oncogenic alleles.
Figure 8. Summary of Validated Rare Functional Alleles of Oncogenes.
Stick plot representation of predicted protein sequence and domain structure of oncogenes harboring rare, functional alleles. Canonical hotspot mutations are shown for reference (gray). Rare variants shown in red scored in eVIP, the EGFR epistasis screen, and the tumor formation screen, and additionally are sensitive to MEK inhibition with trametinib.
DISCUSSION
Here we present a large-scale mapping of lung adenocarcinoma-associated variants to function for 194 alleles representing 53 genes. By focusing on individual variant alleles, we assigned functional significance to alleles that include rare “n of 1” variants. We identified gain-of-function activity for rare variants encoded by the ARAF, BRAF, EGFR, ERBB2, KRAS, and RIT1 oncogenes. Previously, in the absence of functional data, some of these non-canonical variants were excluded from the “known oncogene-positive” set of tumors (TCGA, 2014), but the functional evidence we presented here would support their inclusion as likely driver oncogenes.
The major challenge that this work addresses is that no single assay can rapidly profile the functional impact of a diverse set of genes and alleles (e.g. receptor tyrosine kinases, E3 ligases, transcription factors, small GTPases and more). To tackle this challenge we developed an approach, expression-based variant-impact phenotyping, to infer mutation impact from expression profiling data. eVIP can profile many genes and alleles at once, regardless of function. To validate this approach, we performed two complementary cancer phenotype screens and observed that mutations classified by eVIP as gain-of-function or change-of-function were the only alleles to score in both screens whereas 97% of loss-of-function or neutral alleles failed to score. The concordance between these approaches establishes a proof-of-principle for expression-based variant impact phenotyping and motivates continued systematic functional analyses of genetic variants in cancer.
Application of eVIP to lung adenocarcinoma alleles confirmed the functional impact of canonical and non-canonical gain-of-function oncogenic mutations, but additionally stratified and characterized mutations in genes not read out by the complementary positive-selection studies employed. eVIP analysis revealed a surprisingly high rate of impactful, loss-of-function missense mutations (92%) in the tumor suppressor genes KEAP1 and STK11, indicating that most somatic variation in these genes in lung adenocarcinoma is inactivating, rather than only nonsense and frameshift mutations. However, neutral mutations are also observed, underscoring the importance of continuing to develop and apply computational and functional genomics methods to separate impactful from neutral mutations.
In the present study, we profiled mutations after overexpression in a limited number of lung cellular contexts. We cannot exclude the possibility of false negatives due to context specificity of the activity of individual gene alterations. Therefore, we recognize that these functional studies may not yet be at saturation. At the same time, our studies of rare oncogenic and loss-of-function alleles across cellular contexts were largely concordant and demonstrate that positive results are likely to be robust.
The work presented here demonstrates proof of principle that high-throughput phenotyping of somatic mutations can distinguish impactful mutations from neutral mutations and generate valuable insights into patterns of functional mutations in cancer. These efforts are amenable to extension to include testing of both germline and somatic variants, and the use of genome-editing techniques to study endogenous mutations. Such future efforts might include saturating mutagenesis analysis of a small number of clinically-actionable genes as well as a broad survey of mutated alleles in diverse genes.
Iteration of functional profiling, statistical genomics, and algorithms based on evolutionary and structural constraints will gradually improve the assignment of mutation function. Together, the application of these approaches for classification of mutations will begin to match the pace of genomic discovery and will accelerate the translation of genomic knowledge to clinical care.
EXPERIMENTAL PROCEDURES
Mutated cDNA library
Wild-type open-reading frame constructs (ORFs) were obtained from the human ORFeome library version 5.1 (http://horfdb.dfci.harvard.edu) and used as templates for site-directed mutagenesis to generate mutated cDNAs in the pDONR223 Gateway entry vector. All constructs used in downstream analyses were validated by Sanger sequencing to include the intended mutation and no other identified sequence differences relative to the wild-type construct. After sequence verification, mutated ORFs were shuttled into the pLX317 lentiviral expression vector by LR recombination. All constructs will be publicly available via www.addgene.org.
Cell lines and lentiviral transduction for expression profiling
Cells were plated in 384-well plates and the next day transduced with lentiviral particles carrying ORF constructs. Viral particles were removed 18-24 hours post-infection and cells cultured for 72 hours until L1000 profiling (96 hours total post-transduction). To assess infection efficiency, cells were treated with or without antibiotic selection 24 hours post-infection, and cell viability was determined using CellTiterGlo (Promega) after 72 hours of selection. For the remaining plates, media was removed 96 hours post-infection, and cells were lysed with addition of TCL buffer (Qiagen). Plates were then stored at −80 °C until cDNA synthesis and gene expression profiling.
L1000 profiling and data processing
Luminex bead-based high-throughput gene expression profiling was performed as described in Peck et al., 2006. Standardized processing pipelines of L1000 data developed by the Connectivity Map were used for quantile normalization of expression levels and determination of Z-scored differential gene expression profiles. Z-score calculations were based on plate-wide expression levels. In the case of replicate collapsed profiles, all replicates that passed QC were collapsed based on a weighted average of Z-scores, where the weights correspond to pairwise replicate correlation. More detailed information on standardized data processing and QC can be found on the lincscloud website: http://support.lincscloud.org/. To verify that ORF constructs were adequately expressed, the experiment included 76 cDNAs known to be detected by L1000 probes (“OE CONTROL” in Table S1). Wells containing these expression controls were identified to have the highest upregulation of the respective gene across the dataset in 75 of 76 cases, with a median Z-score of differential expression of 9.4, confirming that most ORFs are robustly expressed as expected.
EGFR epistasis assay
400 PC9 cells were plated per well of 384-well plates and the next day transduced with lentivirus. 48 hours post-transduction, media was changed to fresh media including 300 nM erlotinib, 3 μM erlotinib, or DMSO. 72 hours post-treatment, cell viability was determined using CellTiterGlo reagent (Promega) and luminescence quantified on an Envision MultiLabel Plate Reader (PerkinElmer).
Pooled tumor formation screen
SALE-YAP15SA cells were transduced in 96-well plates, selected with puromycin and expanded, and then pooled. Two million cells per pool per site were injected subcutaneously into immunocompromised mice and tumor formation monitored. Animals were sacrificed when tumor length exceeded 2 cm. Genomic DNA was extracted and subjected to PCR and next-generation sequencing to determine the relative proportion of each barcode within the resulting tumor. All experiments were approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
Supplementary Material
1
2
3
4
5
SIGNIFICANCE.
Variant interpretation remains a major challenge to the implementation of precision medicine. To address this challenge, we developed a gene-expression based method that can characterize the function of many genetic variants, regardless of gene or mutation function. Application of this approach identified 31% of lung adenocarcinoma mutations as neutral, yet 92% of missense variants in tumor suppressor genes KEAP1 and STK11 were identified as loss-of-function variants. Combining this approach with assays for cancer phenotypes identified over a dozen rare, non-canonical mutations as gain-of-function, likely “driver” oncogenic mutations. These data demonstrate the feasibility of systematic functional interpretation of the cancer genome.
HIGHLIGHTS.
- Expression-based phenotyping distinguishes neutral from impactful mutations
- 92% of missense mutations in KEAP1 and STK11 diminish gene function
- Rare variants in ARAF, BRAF, EGFR, ERBB2, KRAS, and RIT1 are oncogenic
- Erlotinib-resistance induced by rare variant mutations is MEK-dependent
ACKNOWLEDGEMENTS
We thank members of the Meyerson and Golub labs for critical advice and discussion. We thank Iris Fung for graphical design of Figure 1. We thank Heidi Greulich and Tanaz Sharifnia for reagents and advice. We are grateful for the feedback and advice from Anne Carpenter, Eejung Kim, Nina Ilic, Lihua Zou, William Kim, Cory Johannessen, Steven Corsello, and William Hahn. The work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation in Mexico, with additional support of National Cancer Institute grant 1R35CA197568 and an American Cancer Society Research Professorship to M.M., an American Cancer Society Postdoctoral Research Fellowship (122398-PF-12-080-01-TBG) and a National Cancer Institute Pathway to Independence award (K99CA197762) to A.H.B. A.N.B was a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2138-12).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, A.H.B, A.N.B., X.W., T.R.G., M.M., and J.B; Methodology, A.H.B, A.N.B., X.W., Y.S., C.C., L.H., F.P., M.B., C.Z., X.Y., A.K., D.R., R.N., I.T., G.G., J.D., P.T., A.S., T.R.G., M.M., and J.B; Software, A.N.B., M.I., L.H., R.S., S.S., A.K., R.N., T.N., D.L.; Validation, A.H.B., X.W., Y.S., and C.C.; Formal Analysis, A.H.B., A.N.B., X.W., Y.S., C.C., F.P., M.B., M.I., L.H., A.K., J.D.C., T.N., D.L., I.T., and J.D.; Investigation, A.H.B, X.W., Y.S., C.C., F.P., M.B., C.Z., X.Y., N.K., S.P., and J.W.; Writing – Original Draft, A.H.B., A.N.B., M.M., J.B.; Writing – Review & Editing, all authors; Visualization, A.H.B, A.N.B., X.W., M.I., R.S., and B.W.; Supervision, T.R.G., M.M. and J.B.; Funding Acquisition, A.H.B, A.N.B., T.R.G., M.M. and J.B.
ACCESSION NUMBERS
The GEO accession number for the L1000 data reported in this paper is GSE83744.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Imielinski M, Duke F, Wala J, Kaplan N, Shi GX, Andres DA, Meyerson M. Oncogenic RIT1 mutations in lung adenocarcinoma. Oncogene. 2014;33:4418–4423. doi: 10.1038/onc.2013.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Cheung HW, Agarwalla PK, Thomas S, Zektser Y, Karst AM, Boehm JS, Weir BA, Berlin AM, Zou L, et al. In vivo multiplexed interrogation of amplified genes identifies GAB2 as an ovarian cancer oncogene. Proc Natl Acad Sci U S A. 2014;111:1102–1107. doi: 10.1073/pnas.1311909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohling S, Scholl C, Levine RL, Loriaux M, Boggon TJ, Bernard OA, Berger R, Dohner H, Dohner K, Ebert BL, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–513. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hast BE, Cloer EW, Goldfarb D, Li H, Siesser PF, Yan F, Walter V, Zheng N, Hayes DN, Major MB. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res. 2014;74:808–817. doi: 10.1158/0008-5472.CAN-13-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Greulich H, Kaplan B, Araujo L, Amann J, Horn L, Schiller J, Villalona-Calero MA, Meyerson M, Carbone DP. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. J Clin Invest. 2014;124:1582–1586. doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpivaara O, Aaltonen LA. Diagnostic cancer genome sequencing and the contribution of germline variants. Science. 2013;339:1559–1562. doi: 10.1126/science.1233899. [DOI] [PubMed] [Google Scholar]
- Kitzman JO, Starita LM, Lo RS, Fields S, Shendure J. Massively parallel single-amino-acid mutagenesis. Nat Methods. 2015;12:203–206. doi: 10.1038/nmeth.3223. 204 p following 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 2014;42:e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, Jackman DM, Johnson BE, Janne PA. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. A method for high-throughput gene expression signature analysis. Genome biology. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Science translational medicine. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, Zhao A, Busuttil RW, Yee H, Stein L, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifnia T, Rusu V, Piccioni F, Bagul M, Imielinski M, Cherniack AD, Pedamallu CS, Wong B, Wilson FH, Garraway LA, et al. Genetic modifiers of EGFR dependence in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2014;111:18661–18666. doi: 10.1073/pnas.1412228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JC, Martin HC, Lise S, Broxholme J, Cazier JB, Rimmer A, Kanapin A, Lunter G, Fiddy S, Allan C, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47:717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich DC, Kryukov G, Carter SL, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson FH, Johannessen CM, Piccioni F, Tamayo P, Kim JW, Van Allen EM, Corsello SM, Capelletti M, Calles A, Butaney M, et al. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell. 2015;27:397–408. doi: 10.1016/j.ccell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5