DNAJC21 Mutations Link a Cancer-Prone Bone Marrow Failure Syndrome to Corruption in 60S Ribosome Subunit Maturation (original) (raw)
Abstract
A substantial number of individuals with bone marrow failure (BMF) present with one or more extra-hematopoietic abnormality. This suggests a constitutional or inherited basis, and yet many of them do not fit the diagnostic criteria of the known BMF syndromes. Through exome sequencing, we have now identified a subgroup of these individuals, defined by germline biallelic mutations in DNAJC21 (DNAJ homolog subfamily C member 21). They present with global BMF, and one individual developed a hematological cancer (acute myeloid leukemia) in childhood. We show that the encoded protein associates with rRNA and plays a highly conserved role in the maturation of the 60S ribosomal subunit. Lymphoblastoid cells obtained from an affected individual exhibit increased sensitivity to the transcriptional inhibitor actinomycin D and reduced amounts of rRNA. Characterization of mutations revealed impairment in interactions with cofactors (PA2G4, HSPA8, and ZNF622) involved in 60S maturation. DNAJC21 deficiency resulted in cytoplasmic accumulation of the 60S nuclear export factor PA2G4, aberrant ribosome profiles, and increased cell death. Collectively, these findings demonstrate that mutations in DNAJC21 cause a cancer-prone BMF syndrome due to corruption of early nuclear rRNA biogenesis and late cytoplasmic maturation of the 60S subunit.
Introduction
Inherited bone marrow failure (BMF) syndromes are a heterogeneous group of life-threatening disorders characterized by a hematopoietic defect in association with a range of variable extra hematopoietic features. Recognized syndromes include Fanconi anemia (MIM: 227650),1 dyskeratosis congenita (MIM: 305000),2 Shwachman Diamond syndrome (MIM: 260400),3 and Diamond Blackfan anemia (MIM: 105650).4 Cells from Fanconi anemia cases exhibit increased chromosomal breakage due to specific genetic and/or functional defects causing genome instability.1 Dyskeratosis congenita cases are characterized by diagnostic mucocutaneous features such as abnormal skin pigmentation, nail dystrophy, and leukoplakia together with the presence of very short telomeres.2 Shwachman Diamond syndrome is characterized by exocrine pancreatic insufficiency, and individuals with Diamond Blackfan anemia are typically reported to have craniofacial abnormalities, including clefting of the lip or palate, thumb abnormalities, cardiac malformations, and short stature.5 Importantly, these syndromes are also associated with an increased cancer risk.
In addition to the well-defined BMF syndromes, we have accrued a number of cases to our BMF registry that do not fit any definite diagnostic classification. These uncharacterized individuals have BMF together with one or more somatic abnormalities, suggesting a constitutional or inherited basis, but do not fulfil the clinical criteria of the known BMF syndromes. With the advent of whole-exome sequencing, it is now possible to unify subsets of these cases through the identification of their underlying genetic defect. This is the case here, as we identify a cohort of simplex BMF cases harboring biallelic mutations in DNAJC21. Through characterization of the encoded protein, we define a cancer-prone BMF syndrome caused by defective DNAJC21.
Material and Methods
Study Approval
All experiments were conducted with the approval of Barts and The London Hospital. Peripheral blood samples were obtained with written consent under the approval of our local research ethics committee (London – City and East).
DNA Sequencing and Plasmids
All samples were obtained with informed written consent and the approval of our local ethics committee. Genomic DNA was extracted from peripheral blood with Gentra reagents (QIAGEN). For exome sequencing, 50 ng of genomic DNA underwent library preparation and exome capture via the Illumina Nextera Rapid Capture Exome kit. Sequencing was performed with the Illumina HiSeq 2000 system, and 100 bp paired-end reads were generated and the data processed through the Illumina pipeline. Variants were called as described previously.6 Resequencing of DNAJC21 was performed as a part of a 32 gene panel and amplified with a TruSeq Custom Amplicon reagent, which was then prepared for MiSeq sequencing according to the manufacturer’s instructions (Illumina). All relevant calls were confirmed by Sanger sequencing. DNAJC21 wild-type and mutant expression constructs were obtained by standard cDNA cloning and mutagenesis procedures into peGFP-C1 (Clontech) plasmid.
Nuclear rRNA Co-immunoprecipitation Assay
Dialyzed nuclear extracts from ∼106 cells expressing eGFP alone or eGFP-tagged wild-type and mutant forms of DNAJC21 were subjected to immunoprecipitation with GFP-TRAP agarose beads (ChromoTek). After incubation and stringent washes, immune complexes were treated with DNase I (RNase-free) (Fermentas) for 30 min at 37°C in the presence of RNasein (Invitrogen). The reactions were stopped by addition of 2.5 mM EDTA and heating for 10 min at 65°C. Co-immunoprecipitated RNA was extracted by Trizol (Invitrogen), purified with RNeasy kit columns (QIAGEN), and analyzed by qRT-PCR for precursor 45S rRNA using primers indicated in Table S3.
qRT-PCR
RNA from nuclear extracts was purified with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed with SuperScript III Reverse Transcriptase (Invitrogen). qPCR was carried out on a real-time PCR detection system with SYBR Green (Thermo Fisher) for 45S precursor rRNA, mature 28S, and 18S rRNA in individual 4, both parents, and three unrelated control individuals. Data presented is normalized to GAPDH mRNA amounts, and expression of SNORA63 and SNORA68 was determined as internal standard controls.
RNAi Studies
All siRNA and shRNA experiments were performed on HeLa cells cultured at 37°C in 5% CO2 and DMEM (Cambrex), supplemented with L-glutamine, 10% fetal calf serum (FCS), and penicillin and/or streptomycin antibiotics. Transfections of DNAJC21 siRNA (Sigma, cat. no. EHU103331) were carried out with 50 nM siRNA and Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocol; a predesigned non-target siRNA (Ambion) was used as a negative control. Cells were harvested at 72 hr for subsequent analysis. Tet-pLKO-puro shRNA-expressing plasmids were obtained from Addgene and prepared using a protocol adopted from Addgene7 to generate Tet-pLKO-puro DNAJC21 3′ UTR shRNA-specific clones. The specific shRNA sequences of the DNAJC21 3′ UTR used in this study were obtained from the RNAi Consortium (TRC MISSION shRNA library) and the RNAi designer tool from Thermo Fisher. Target sequences are given in Figure S4A. DNAJC21 3′ UTR shRNA expression was induced by culturing cells in DMEM supplemented with tetracycline (Tet)-free serum (Clontech) for 72 hr in the presence of the indicated concentrations of doxycycline (dox; Sigma, Figure S4B) dissolved in deionized water.
Immunostudies
For co-immunoprecipitation studies, approximately 250 μg cell lysate was used per immunoprecipitation in cells transfected with eGFP encoding wild-type and mutant DNAJC21 plasmids. In brief, 72 hr after transfection, lysates were precleared with rabbit control IgG (monoclonal SP137, Abcam) and further incubated with 10 μl anti-GFP antibody (Abcam, cat. no. EPR14104) overnight at 4°C. For determination of nucleic-acid-mediated interaction, cell lysates were initially treated with 40 U DNase I (RNase-free) or 25 μg/mL RNase A for 30 min at 37°C. The next day, 20 μl Protein A Dynabeads (Thermo Fisher) pre-equilibrated with immunoprecipitation (IP) buffer (25 mM Tris-HCl [pH 7.9], 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT, 1× Complete EDTA-free protease inhibitor cocktail) containing 100 mM KCl was added and gently mixed for 2 hr at 4°C. Immune complexes were washed three times with IP buffer containing 150 mM KCl and two times with IP buffer containing 100 mM KCl, each for 5 min at 4°C. Immunoprecipitates were boiled in SDS-sample buffer (25 mM TrisHCl [pH 6.8], 2% SDS, 10% glycerol, 0.05% bromophenol blue, 5% 2-mercaptoethanol) for 5 min, separated by SDS- PAGE, transferred to polyvinylidene membranes, and subjected to western blot analysis with a WesternBreeze Chromogenic kit (Thermo Fisher, cat. no. WB7106 and WB7104). Antibodies against DNAJC21 (Abcam, cat. no. ab86434, C terminal epitope corresponding to amino acids 400–450; Proteintech, cat. no. 23411-1-AP, N terminal epitope corresponding to amino acids 43–288), PA2G4 (Abcam, cat. no. ab119037), ZNF622 (Abcam, cat. no. Ab57859), and HSPA8 (Abcam, cat. no. EP1513Y) were used in the study. For immunocytochemical staining, HeLa cells and IMR-90 cells (primary lung fibroblasts obtained from ATCC CCL-186) were grown on coverslips, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 (TX100) in PBS, quenched in 50 mM NH4Cl, and blocked in 10% goat serum and 1% BSA in PBS containing 0.05% TX100 for 1 hr. Cells were incubated with corresponding primary and secondary antibodies and mounted via vectashield containing DAPI (Vector Labs, cat. no. H-1200). Images were collected with an LSM710 laser scanning confocal microscope (Olympus) under relevant excitation, and the emitted signals were visualized with ZEN software (Zeiss). Nucleolar co-staining was determined by mouse monoclonal antibody against nucleophosmin (Abcam, cat. no. ab40696) and DNAJC21.
Sub-cellular Fractionation
DNAJC21 knockdown and control cells were lysed in ice-cold HEPES containing 0.1% NP40 and centrifuged at 3,000 rpm, and the supernatant was collected for extraction of cytoplasmic protein. The pellet-containing nuclei were briefly washed three times in ice-cold HEPES containing 0.1% NP40, were added to the pellet ice-cold radioimmunoprecipitation assay (RIPA) buffer containing a cocktail of protease and phosphatase inhibitors (Roche, cat. no. 04693116001), and were sonicated twice for 10 s at 50% pulse to release nuclear proteins. The final mixture was shaken gently on ice for 15 min, and the nuclear fraction protein supernatant was obtained by centrifugation at 14,000 g for 15 min. Fractionated lysates were verified with antibodies against cytoplasmic GAPDH (Abcam, cat. no. EPR16891) and nuclear TATA binding protein (Abcam, cat. no. EPR3826) by immunoblotting as described above.
T Cell Isolation and Culture
Blood samples were obtained from individual 4 and her unaffected heterozygous parents. Peripheral blood mononuclear cells were separated with lymphocyte separation medium (Lonza) according to the manufacturer’s instructions, washed twice with complete RPMI medium containing 15% FCS, and resuspended at 1·0 × 106 cells/mL in RPMI-1640 medium and 20% FCS (Invitrogen). T cell mitosis was stimulated by the addition of 10 μg/mL of phytohaemagglutinin (PHA; Roche Applied Science). After 48 hr, the medium was replaced with fresh RPMI medium containing 10 units/mL of recombinant human interleukin-2 (Invitrogen) and harvested after 14 days. Cells were counted on a NucleoCounter YC-100 automated cell counter (ChaemoMetec). Epstein-Barr virus (EBV)-infected lymphoblastoid cells (LCLs) were established and grown in RPMI-1640 medium supplemented with penicillin and streptomycin, 2 mM L-glutamine, and 20% (vol/vol) FBS (Invitrogen).
Polysome Analysis
For analysis of monosomes and polysomes, HeLa cells and LCLs were grown to confluence, harvested 10 min after treatment with 100 mg/mL cycloheximide (CHX; Sigma), and resuspended in PBS containing 100 μg/mL CHX. Equal numbers of cells were lysed in 425 μL of hypotonic buffer (5 mM Tris-HCl [pH 7.5], 2.5 mM MgCl2, 1.5 mM KCl, and 1× protease inhibitor cocktail [EDTA-free]), with 5 μL of 100 μg/mL CHX, 1 μL of 1 M DTT, and 100 U of RNase inhibitor. Lysates were vortexed for 5 s, followed by addition of 25 μl of 10% Triton X-100 (final concentration 0.5%) and 25 μL of 10% sodium deoxycholate (final concentration 0.5%), and vortexed again for 5 s. Lysates were then centrifuged at 16,000 g for 2 min, and supernatants were layered on 15% to 45% (wt/vol) sucrose gradients. The gradients were centrifuged at 36,000 rpm for 4 hr at 4°C in a Beckman SW41Ti rotor. The fractionated lysates were serially collected, and polysome profile peaks were obtained by measuring absorbance at 254 nm. Proteins from these fractions were precipitated with 20% (vol/vol) trichloroacetic acid, separated on SDS–PAGE gels, and transferred to polyvinylidene membranes for immunoblotting.
Cell Death and Rescue by Wild-Type DNAJC21
After 72 hr of RNAi treatment, cells were washed in PBS; propidium iodide (PI) was added to the cell suspension to a final concentration of 5 μg/mL and analyzed on an LSRII Flow Cytometer (BD Biosciences). Plasmids encoding WT-eGFPDNAJC21 and peGFP-C1 alone were transfected into DNAJC21 3′ UTR shRNA expressing stable cell lines. 48 hr after transfection, cells were fluorescence-activated cell (FAC) sorted by gating for eGFP, collected, and re-plated in 96 wells in equal number. It is important to note that the shRNA system targets the 3′ UTR of the endogenous DNAJC21 mRNA. This UTR is absent from the plasmid-derived transcript, resulting in shRNA depletion of the endogenous DNAJC21 only upon dox induction. DNAJC21 3′ UTR shRNA 1 (Figure S3) was preferred because it showed DNAJC21 knockdown upon dox induction at a lower concentration, 100 ng/mL.
Structural Analysis of the DNAJC21 p.Pro32Ala Variant
The J domain crystal structure (PBD: 1FAF) was obtained from the Protein Data Bank and visualized with Swiss PDB Viewer.8 The predicted effects of the DNAJC21 p.Pro32Ala substitution were generated with this program.
Statistics
Analyses were performed with GraphPad Prism 5.0 software (GraphPad Software). Mann-Whitney U test and one-way ANOVA with Tukey’s post hoc tests were used when appropriate, as indicated within the text or figure legends. Data are presented as the means ± SEM unless otherwise indicated. A two-tailed p value of <0.05 was considered significant.
Results
Biallelic Mutations in DNAJC21 Cause a BMF Syndrome
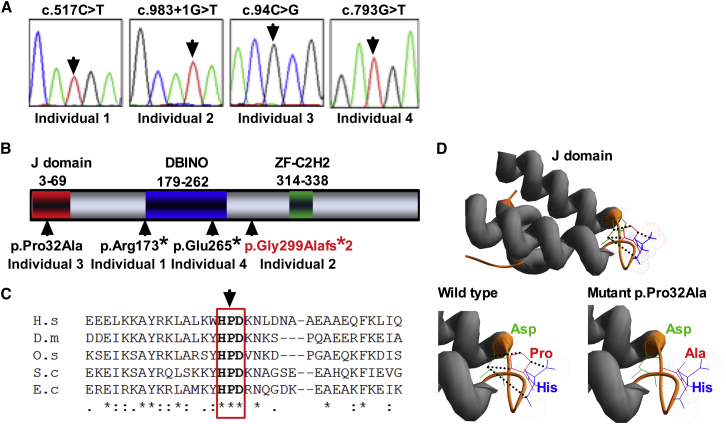
In a cohort of 28 unrelated individuals with BMF and non-specific somatic features from our BMF registry (Table S1), we sought an underlying genetic basis through exome sequencing. Three of these individuals, none of whom had any family history of disease, were found to have homozygous likely pathogenic variants in DNAJC21 ([GenBank: NM_001012339.2] Table 1, Figures 1A and 1B). Specifically, these were a nonsense variant, c.517C>T, p.Arg173∗, in individual 1, a splice variant, c.983+1G>T, in individual 2, and a missense variant, c.94C>G, p.Pro32Ala, in indivdual 3. Targeted resequencing of DNAJC21 in a second cohort of 23 similar individuals (Table S2) identified one further individual (indivdual 4) with a homozygous nonsense variant, c.793G>T, p.Glu265∗ (Table 1). Sanger sequencing of parental DNA revealed that this variant segregated as an autosomal-recessive trait. The four individuals with biallelic DNAJC21 mutations are similar in clinical presentation (Table 1): they have global BMF associated with peripheral pancytopenia and are characterized by intrauterine growth restriction and/or short stature. One of them (individual 3) developed acute myeloid leukemia, sub-type megakaryocytic (AML-M7), at the age of 12 years (Table 1).
Table 1.
Features of Subjects with Biallelic Mutations in DNAJC21
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | |
|---|---|---|---|---|
| Genotype (c.) | 517C>T | 983+1G>T | 94C>G | 793G>T |
| Protein (p.) | Arg173∗ | Gly299Alafs∗2a | Pro32Ala | Glu265∗ |
| Gender | F | F | M | F |
| Age when sample received (years) | 3 | 6 | 12 | 6 |
| Country of origin | France | Algeria | Pakistan | Pakistan |
| Are parents first cousins? | Y | Y | Y | N |
| Bone marrow failureb | Y | Y | Y | Y |
| IUGR and/or short stature | Y | Y | Yc | Y |
| Microcephaly | Y | Y | N | N |
| Other features | N | Yd | Ye | Yf |
| Chromosomal breakageg | N | N | N | N |
| Telomere lengthh | normal | normal | normal | normal |
Figure 1.
Biallelic Mutations in DNAJC21
(A) Sanger sequencing traces and the genotype of each individual are given. Arrows indicate the mutated nucleotides.
(B) The positions of the alterations caused by the mutations observed in our individuals are shown in the context of the conserved functional domains predicted in the DNAJC21 amino acid sequence. Residue numbers are given for the different domains. J domain refers to the DNAJ domain that defines this group of proteins; DBINO refers to the DNA-binding domain found on global transcription activator SNF2L1 proteins and chromatin re-modeling proteins. The frameshift p.Gly299Alafs∗2 variant is shown in red as it is predicted to arise from the deletion of exon 7 due to the splice-site mutation c.983+1G>T in individual 2.
(C) Conservation of the HPD motif in the J domain of DNAJC21. Alignment of residues 16–50 of the human DNAJC21 J domain with four other DnaJ proteins was generated with ClustalW. The arrow indicates the proline residue that was mutated in individual 3. The aligned sequences are human (H.s.) DNAJC21 (UniProt: Q5F1R6), Drosophila melanogaster (D.m.) DNAJ-1 (UniProt: Q53ZT0), rice (O.s.) Oryza sativa DNAJ homolog (UniProt: Q948S9), Saccharomyces cerevisiae (S.c.) DnaJ-related protein SCJ1 (UniProt: P25303), and Escherichia coli (E.c.) chaperone protein DnaJ (UniProt: P08622). Asterisks indicate positions that have a single fully conserved residue, colons indicate conservation between groups of strongly similar properties, and periods indicate conservation between groups of weakly similar properties.
(D) E. coli DnaJ domain topology (PDB:1FAF) visualized with Swiss PDB Viewer. Ribbon diagram depicts J domain (gray) and position of HPD motif (orange loop). The hydrogen bonds between residues in the HDP motif are indicated by dotted lines (black). Mimicking the individual 3 mutant by in silico analysis revealed loss of the proline’s cyclic ring, disrupting hydrogen bonds (dotted line, black), when substituted with alanine in the J domain.
Three of the variants are predicted to cause loss of function (LOF, defined here as nonsense or splice variants), and the transcripts containing these nonsense and predicted frameshift mutations are likely to undergo nonsense-mediated mRNA decay. In the fourth individual, a missense variant (p.Pro32Ala) disrupts the highly conserved histidine-proline-aspartic acid (HPD) motif that lies at the heart of the J domain, which defines a family of proteins (Figures 1B and 1C).10 In silico analysis of the p.Pro32Ala substitution on the tertiary structure of the E. coli J domain (PDB: 1FAF) reveals the loss of the distinctive cyclic structure of proline in a loop between two α helices (Figure 1D). This structural alteration to the HPD motif is likely to disrupt the interaction of the J domain with its cognate heat shock protein 70 and the subsequent stimulation of ATPase.11 To our knowledge, none of the variants have been reported previously apart from the splice variant (c.983+1 G>A), which is present only in the heterozygous state in the Exome Aggregation Consortium (ExAC) database at an allele frequency of 5/120,170.
The ExAC database estimates the frequency of LOF alleles in DNAJC21 (combining frameshift, nonsense, and splice donor- and acceptor-site variants) at about 0.1% (123 LOF alleles in ∼120,000 chromosomes). Hence, the frequency of LOF homozygotes and compound heterozygotes is expected to be in the range of one per million individuals. Not including the likely damaging homozygous missense variant, the observed frequency of LOF homozygous cases in our combined cohort (3 out of 51 individuals) significantly exceeds this expectation (binomial test p < 10−10). We conclude that the allelic series of mutations that we have identified here defines a subgroup of individuals with constitutional BMF caused by defective DNAJC21.
DNAJC21 Is Implicated in rRNA Biogenesis
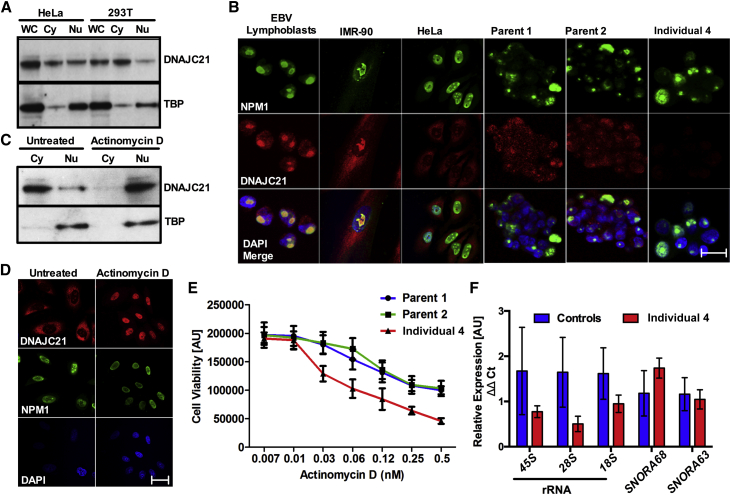
DNAJC21 belongs to the family of DnaJ (heat shock protein 40) chaperone proteins11 and is ubiquitously expressed in human tissues (Figure S1). We found that DNAJC21 was present in both cytoplasm and the nucleus of HeLa and 293T cells. (Figure 2A) Within the nucleus, it localized primarily to the nucleolus in three different cell lines tested (Figure 2B). T lymphocytes obtained from one affected individual (individual 4) revealed a lack of DNAJC21 immunoreactivity to an antibody recognizing an N terminal epitope, whereas the parental control T cells stained positive (Figure 2B).
Figure 2.
Nucleolar Localization of DNAJC21 and Its Role in rRNA Biogenesis
(A) DNAJC21 is present in both cytoplasmic (Cy) and nuclear fractions (Nu). Antibody against TBP was used as a control for nuclear fractions. WC, whole cell.
(B) Immunocytochemistry shows the subcellular localization of DNAJC21 in different cell types. A lack of DNAJC21 immunostaining is observed in T cells from individual 4, whereas the parental T cells stain positive. Nucleophosmin (NPM1) is used as a control. Images display NPM1 (green), DNAJC21 (red), and DAPI (blue). Scale bar, 20 μm.
(C and D) DNAJC21 translocates to the nucleus after actinomycin D treatment. NPM1 is used as positive control.
(E) LCLs from individual 4 and asymptomatic heterozygous parent controls (1 and 2) were plated in the presence of increasing concentrations of actinomycin D for 48 hr and assayed for cell viability by staining with neutral red. Assays were performed in octuplets per experiment and repeated for a minimum of two independent experiments. Data points represent mean ± SEM.
(F) Individual 4 LCLs show reduced expression for rRNA in nuclear extracts analyzed when compared to both those of parents and three unrelated samples as controls. Expression of SNORA63 and SNORA68 was determined as internal controls. All genes are normalized relative to expression of GAPDH mRNA. Data represent mean ± SD, n = 2, performed in triplicates.
To date, little is known about human DNAJC21 apart from a report that its depletion causes a “miscellaneous” effect on rRNA processing in HeLa cells.12 To investigate the possible relationship between DNAJC21 and rRNA, HeLa cells were treated with low levels (2 nM) of actinomycin D (ActD) that are sufficient to exert its inhibitory effect on rRNA synthesis.13 This treatment resulted in a translocation of DNAJC21 from the cytoplasm to the nucleus (Figures 2C and 2D). We also observed that LCLs derived from individual 4 showed an increased sensitivity to ActD treatment when compared to parent controls (Figure 2E). qRT-PCR analysis of nuclear RNA extracts obtained from these cells showed lower rRNA levels in the affected individual than in both parents and unrelated control individuals (Figure 2F). These results indicate that DNAJC21 is involved in rRNA biogenesis.
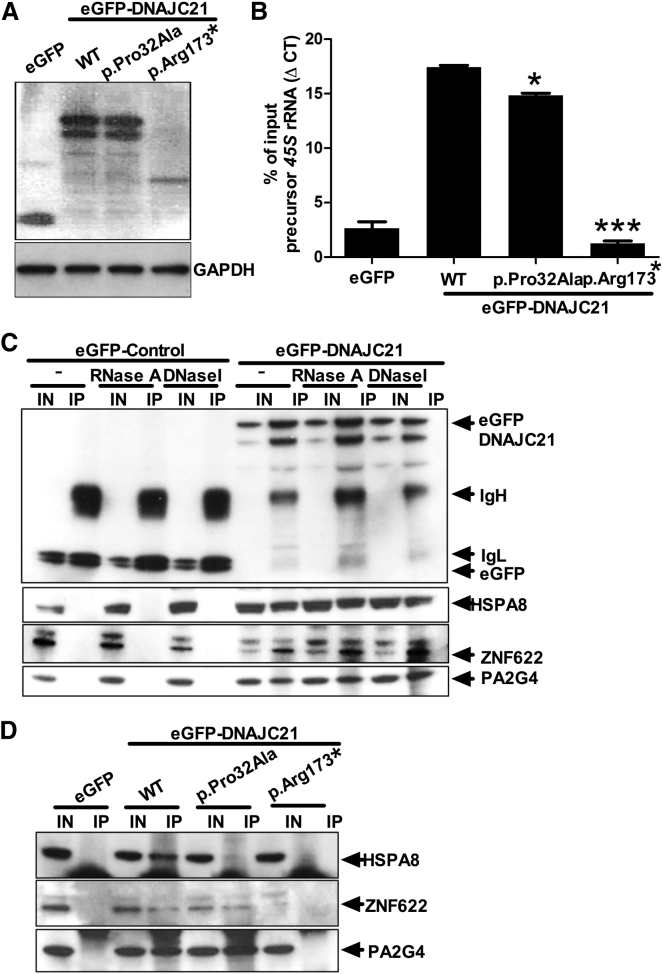
Given its nucleolar localization and response to ActD, we sought to investigate a role for DNAJC21 in 45S precursor rRNA binding. We therefore performed native RNA co-immunoprecipitation experiments on nuclear extracts from HeLa cells expressing wild-type and mutant forms of GFP-tagged DNAJC21. Amounts of ectopically expressed GFP-tagged proteins were established by immunoblotting with an anti-GFP antibody (Figure 3A). qRT-PCR analysis on GFP-TRAP immunoprecipitates showed that DNAJC21 associates with the precursor 45S rRNA. The truncated DNAJC21 mutant (p.Arg173∗) failed to bind to precursor 45S rRNA (Figure 3B). Taken together, these results support the notion that DNAJC21 is indeed a pre-rRNA processing factor,12 given that it associates with precursor 45S rRNA and is involved in rRNA biogenesis.
Figure 3.
DNAJC21 Associates with rRNA and Interacts with 60S Ribosome Maturation Factors
(A) Expression of eGFP-tagged DNAJC21 wild-type and mutants was detected by immunoblotting with anti-GFP. GAPDH was used as a loading control.
(B) Immunoprecipitation was performed with GFP-TRAP agarose beads and nuclear extracts of HeLa cells expressing wild-type and mutant forms of DNAJC21. Cells expressing EGFP alone were used as a control for the co-immunoprecipitation. RNA was extracted from the immunoprecipitates and analyzed by RT-PCR. Data represent mean ± SD, n = 2 independent experiments performed in duplicate. ∗p < 0.05, ∗∗∗p < 0.0001 Mann-Whitney U test.
(C) Co-immunoprecipitations were performed in HeLa cells expressing eGFP or eGFP-DNAJC21. Lysates were treated with DNase I or RNase A prior to immunoprecipitation where indicated. Co-immunoprecipitated proteins were detected by western blot analysis with eGFP-, HSPA8-, ZNF622-, and PA2G4-specific antibodies.
(D) Co-immunopreciptation from cells expressing eGFP-tagged wild-type and mutant forms of DNAJC21 reveal the variable interaction with cofactors involved in 60S ribosome maturation. IN, 10% input; IP, immunoprecipitate; WT, wild-type.
DNAJC21 Interacts with 60S Ribosome Maturation Factors
The yeast ortholog of DNAJC21, Jjj1 (Figure S2A), has been studied in some detail. In addition to its role in rRNA processing in the nucleus,14 Jjj1 has been shown to act in the final stages of 60S ribosome subunit maturation in the cytoplasm.14, 15, 16, 17, 18, 19 Specifically, Jjj1 is responsible for the eviction of the 60S nuclear export receptor, Arx1 (human orthologs, proliferation-associated protein 2G4 [PA2G4], also known as Erbb3-binding protein 1 [EBP1], and IRES-specific cellular transacting factor 45 [ITAF45], Figure S2B).20, 21, 22 In order to execute this function, Jjj1 acts in concert with Rei1 (human orthologs, zinc finger protein 622 [ZNF622], Figure S2C), binding to the 60S subunit. Together, Jjj1 and Rei1 recruit Ssa1p (human ortholog, heat shock 70 kDa protein 8 [HSPA8], Figure S2D) to stimulate ATPase activity. This takes place in the cytoplasm, allowing Arx1 to recycle back into the nucleus to aid in further rounds of 60S nuclear export (Figure S3).
To determine whether these interactions also occurr in humans, we expressed eGFP-tagged wild-type and mutant forms of DNAJC21 in HeLa cells. Co-immunopreciptation experiments revealed the predicted interactions of DNAJC21 with PA2G4, ZNF622, and HSPA8 (Figure 3C). These interactions were not mediated by nucleic acids given that they were detected in both DNase-I- and RNase-A-treated cell lysates (Figure 3C). In contrast, expression of the truncation mutant (p.Arg173∗) revealed no detectable interaction with any of the aforementioned proteins (Figure 3D). We note that it is likely that in vivo, the transcript encoding p.Arg173∗ undergoes nonsense-mediated decay, and the variant is included in these studies as a negative control. The missense variant (p.Pro32Ala) did interact with PA2G4 and ZNF622, but failed to interact with HSPA8 (Figure 3D). This is consistent with studies in yeast, which have shown that mutation of the HPD motif in Jjj1 results in loss of interaction with its HSP70 binding partner, Ssa1p.14, 16, 18
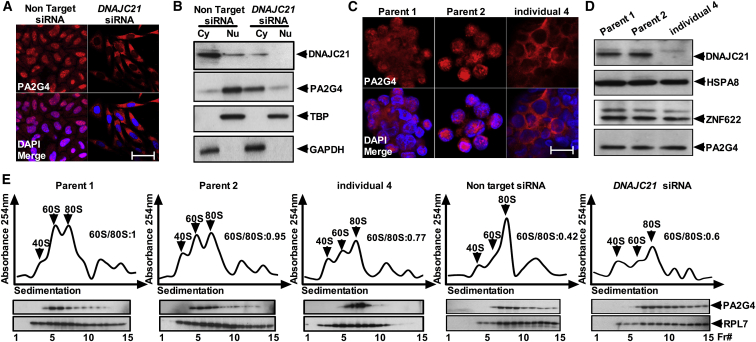
Because these mutants failed to interact with 60S maturation factors, we further investigated the role of DNAJC21 in ribosome biogenesis. Immunostudies revealed a dramatic accumulation of PA2G4 in the cytoplasm of _DNAJC21_-knockdown cells as well as T lymphocytes from individual 4, in comparison to the predominantly nuclear localization observed in the relevant controls (Figures 4A–4C). These data suggest that loss of DNAJC21 perturbs PA2G4 traffic and is consistent with previous studies in yeast, which showed that Arx1 fails to traffic back to the nucleus in strains that lack Jjj1.15, 17, 18, 19 However, there was no change in the relative abundance of PA2G4, ZNF622, and HSPA8 in individual 4 LCLs compared to those of parent controls (Figure 4D). Sucrose density centrifugation analysis revealed abnormal polysome profiles in individual 4 LCLs, as well as in _DNAJC21_-knockdown cells, in comparison to controls (Figure 4E). Furthermore, a stark increase in the amount of PA2G4 bound to 60S and 80S peak fractions is observed in individual 4 LCLs (Figure 4E). Collectively, these observations indicate that human DNAJC21 plays a prominent role in the maturation of the 60S ribosome subunit.
Figure 4.
Loss of DNAJC21 Impairs Traffic of PA2G4, Corrupting Ribosome Biogenesis
(A–C) Cytoplasmic accumulation of PA2G4 was observed in DNAJC21-knockdown cells and T lymphocytes from individual 4 when compared to relevant controls. (A) and (C) show DAPI (blue) and PA2G4 (red). Scale bars, 35 μm (A) and 20 μm (C). Panels indicate representative images taken from different fields of view in three separate experiments. (B) shows immunoblotting of cytoplasmic (C) and nuclear (N) fractions.
(D) Immunoblotting of LCL lysates from individual 4 and parental controls for DNAJC21 and 60S maturation factors HSPA8, PA2G4, and ZNF622.
(E) Cycloheximide-treated lysates from individual 4 and parent control LCLs, as well as DNAJC21 and non-target siRNA-treated HeLa cells, were fractionated on 15% to 45% sucrose gradients by ultracentrifugation. Absorbance at 254 nM across the gradient fraction is shown (top panels). Ribosomal subunit ratios (60S:80S) are indicated. TCA-precipitated proteins from equal aliquots of each fraction were immunoblotted for PA2G4 and RPL7 as a marker for 60S and 80S subunits and polysomes. Data presented is representative of two independent experiments (n = 2)
Loss of DNAJC21 Disrupts Cell Morphology and Inhibits Cell Growth
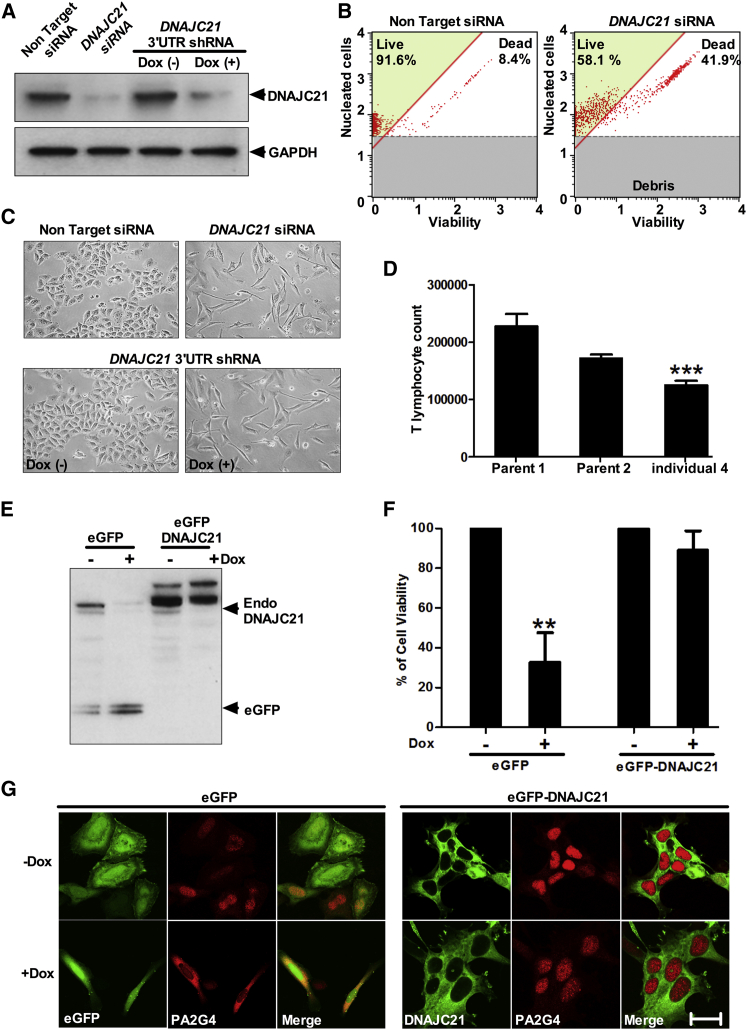
Defective ribosome biogenesis could disrupt protein translation, impairing cellular growth. In line with this, knockdown of DNAJC21 induced cell death in HeLa cells (Figures 5A and 5B). DNAJC21-knockdown cells also exhibited an unusual morphology with markedly elongated cellular morphology (Figure 5C). T cells obtained from individual 4 also showed a significant impairment in growth rate after mitogenic stimulation with phytohemagglutinin and interleukin-2, relative to parental controls (Figure 5D, p < 0.001, Student’s t test).
Figure 5.
Loss of DNAJC21 Inhibits Cell Growth
(A) After 72 hr of DNAJC21 siRNA by doxycycline (dox) treatment to induce shRNA targeting the 3′ UTR of DNAJC21 (100 ng/mL), whole-cell extracts were subjected to western blot analysis with DNAJC21- and GAPDH-specific antibodies.
(B) FAC sorting plots show increased cell death in cells treated with DNAJC21 siRNA.
(C) Phase contrast microscopy images of _DNAJC21_-RNAi-treated cells exhibiting abnormal morphology in comparison to controls. All panels are representative of images taken from different fields of view in two separate experiments.
(D) T lymphocytes from individual 4 show impaired growth as compared to parental controls. Data represent mean ± SEM; ∗∗∗p < 0.001, Student’s t test.
(E) Knockdown of endogenous DNAJC21 by dox administration is detected by western blotting using a mix of both DNAJC21 and GFP antibodies in stably transduced DNAJC21 3′ UTR shRNA HeLa cells. Arrows indicate positions of endogenous DNAJC21 and eGFP control.
(F) Cell viability relative to an untreated dox (−) culture was measured by GFP fluorescence. This revealed the rescue of cell viability in the presence of eGFP-DNAJC21 upon dox induction of the shRNA targeting the endogenous transcript. Data represent mean ± SEM; ∗∗p < 0.01 one-way ANOVA with Tukey’s post hoc test.
(G) In cells expressing eGFP alone, confocal imaging revealed cytoplasmic accumulation of PA2G4 and abnormal morphology upon dox administration. However, in cells expressing eGFP-DNAJC21, no defect in PA2G4 shuttling to the nucleus or cell morphology was observed upon dox addition (+) when compared to an untreated dox (−) culture. Images display eGFP (green) and PA2G4 (red). Panels are representative of images taken from different fields of view in three separate experiments. Scale bar, 20 μm.
To confirm that the cell death, cytoplasmic accumulation of PA2G4, and altered morphology were a consequence of DNAJC21 deficit, we performed experiments using a dox-inducible shRNA system that targeted the 3′ UTR of DNAJC21. Cell lines expressing DNAJC21 3′ UTR shRNA were transfected with either eGFP-DNAJC21 or the eGFP vector alone. Knockdown of the endogenous protein and the ectopic expression of eGFP-tagged wild-type DNAJC21 (which escapes the effect of the shRNA) were verified by immunoblotting (Figure 5E). As expected, re-introduction of wild-type DNAJC21 rescued cell viability and restored normal PA2G4 traffic and morphology (Figures 5F and 5G), demonstrating that the observed effects were a specific consequence of DNAJC21 depletion.
Discussion
Although the inherited BMF syndromes have several distinguishing hallmarks, their heterogeneous presentation and overlapping features have often confounded their clinical diagnosis. However, as the underlying disease associated genes have been discovered, our understanding of BMF pathogenesis has improved considerably. Now, in the era of next-generation sequencing, it is possible to identify subgroups of cases, unified by their underlying genetic defects. In this study, we have used whole-exome sequencing to identify a distinct subgroup of individuals presenting with constitutional BMF characterized by defective DNAJC21.
We have shown that DNAJC21 associates with precursor 45S rRNA. Cells from affected individuals harboring biallelic DNAJC21 mutations had reduced amounts of precursor 45S and mature 28S rRNA and exhibited increased sensitivity to ActD, implicating its role in rRNA biogenesis. In addition, DNAJC21 participates in coordinating nucleo-cytoplasmic shuttling of PA2G4, to aid in nuclear export of the 60S subunit. Our findings from DNAJC21 RNAi studies in HeLa cells and cells from an affected individual provide compelling evidence in support of the hypothesis that loss of DNAJC21 perturbs an essential late cytoplasmic step of the 60S subunit maturation, thereby corrupting ribosome biogenesis. Previous studies reported that PA2G4 binds to FG-repeat nucleoporins through its methionine amino peptidase activity and aid in 60S nuclear export.23 In eukaryotic cells, PA2G4 is shown to regulate diverse functions such as cell growth and differentiation, interaction with transcription factors,22 ribosome biogenesis, and rRNA processing20, 24 as well as IRES-mediated translation.21 Based on these reports, it is conceivable that cytoplasmic accumulation of PA2G4 in absence of DNAJC21 would affect this wide range of diverse functions in human cells. Together, our data clearly establish that the function of DNAJC21 in ribosome biogenesis is highly conserved from yeast to humans.14, 15, 16, 17, 18, 19
Altered or reduced ribosome biogenesis has long been implicated in BMF syndromes.5 We now add DNAJC21 to the growing list of ribosomopathies, which are a class of disease caused by mutations that affect the biosynthesis and or function of the ribosome.25 Ribosomopathies are intriguingly enigmatic given that they initially present with pathognomonic features of too few cells in the marrow but can (in ∼10%–20% of cases) later give rise to hyper-proliferative disorders such as acute myeloid leukemia.5, 26 It is notable that this holds true for one of the four individuals presented here, although with such a small number of affected individuals, it is difficult to extrapolate this proportion to a larger cohort. Although it is likely that these cancers arise from a selection pressure resulting in the acquisition of mutations, we also believe that a dearth of the 60S ribosome subunit could exert pressure on cells to use defective ribosomes. Over time, these cells might become error prone, leading eventually to transition from a hypo- to a hyper-proliferative phenotype.26, 27
We have identified two distinct areas of operation of DNAJC21, one in nucleolar rRNA biogenesis and a second in cytoplasmic recycling of nuclear export factor PA2G4 for 60S subunit maturation. We note that the yeast ortholog of human DNAJC21 (Jjj1) acts just upstream of the release of Tif6 by Sdo1 in 60S maturation28 and that the human ortholog of Sdo1 is SBDS, the protein defective in Shwachman Diamond syndrome.29, 30, 31, 32 By analogy with SBDS, it is therefore more likely that the disease phenotype in our individuals is caused by the late 60S maturation deficit. This is also consistent with the suggestion that mutations that affect the overall structural integrity of the ribosome are less likely to be tolerated.27
It is striking that two out of the four individuals presented here have homozygous nonsense variants and we were unable to detect DNAJC21 in the lymphocytes of an affected individual. It therefore appears that individuals who lack DNAJC21 remain viable. This is in contrast to Shwachman Diamond syndrome cases, where no homozygotes for a recurrent nonsense variant have been reported, suggesting that null mutations in SBDS are embryonic lethal.33 We do note, however, that the small numbers of DNAJC21 cases presented here have global BMF by the age of 12 years, whereas in Shwachman Diamond syndrome, the BMF is more variable and often only involves an isolated neutropenia.34
In conclusion, our study defines a cancer-prone BMF syndrome caused by mutations in DNAJC21. Characterization of the encoded protein shows that it plays a highly conserved role in ribosome biogenesis. We have therefore identified a distinct ribosomopathy, highlighting the significance of corrupted ribosomes in the etiology of both hypo-proliferative and hyper-proliferative disorders.
Acknowledgments
We would like to thank all the families and clinicians who contributed to this research. This work was funded by The Medical Research Council UK (MR/K000292/1), Children with Cancer UK (2013/144), Barts and The London Charity (845/1796), and Leukaemia Lymphoma Research/Bloodwise (14032). The authors would like to thank Frederic Baleydier for providing case samples, Gary Warnes for support in FACS analysis, and staff at the Barts and The London Genome Centre for Sanger sequencing analysis.
Published: June 23, 2016
Footnotes
Accession Numbers
Variants identified in DNAJC21 were deposited under accession numbers ClinVar: SCV000257530, SCV000257531, SCV000257532, and SCV000257533.
Web Resources
- 1000 Genomes, http://www.1000genomes.org
- Addgene shRNA Production, http://www.addgene.org/static/data/41/67/165920fc-af64-11e0-90fe-003048dd6500.pdf
- BioGPS, http://biogps.org/
- ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- ExAC Browser, accessed January 2016, http://exac.broadinstitute.org/
- GenBank, http://www.ncbi.nlm.nih.gov/genbank/
- GTEx Portal, http://www.gtexportal.org/home/
- NCBI Conserved Domains, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- OMIM, http://www.omim.org/
- RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
- RNAi Designer, https://rnaidesigner.thermofisher.com/rnaiexpress/setOption.do?designOption=sirna
- Swiss PDB Viewer, http://spdbv.vital-it.ch/
- The RNAi Consortium shRNA Library, https://www.broadinstitute.org/rnai/trc/lib
- UniProt, http://www.uniprot.org/
Supplemental Data
Document S1. Figures S1–S4 and Tables S1–S3
Document S2. Article plus Supplemental Data
References
- 1.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dokal I. Dyskeratosis congenita. Hematology Am. Soc. Hematol. Educ. Program. 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- 3.Shimamura A. Shwachman-Diamond syndrome. Semin. Hematol. 2006;43:178–188. doi: 10.1053/j.seminhematol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Vlachos A., Ball S., Dahl N., Alter B.P., Sheth S., Ramenghi U., Meerpohl J., Karlsson S., Liu J.M., Leblanc T., Participants of Sixth Annual Daniella Maria Arturi International Consensus Conference Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br. J. Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggero D., Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124:2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tummala H., Walne A., Collopy L., Cardoso S., de la Fuente J., Lawson S., Powell J., Cooper N., Foster A., Mohammed S. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015;125:2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y.M., Lengauer C. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 8.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 9.Cawthon R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai J., Douglas M.G. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 11.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Iapalucci-Espinoza S., Franze-Fernández M.T. Effect of protein synthesis inhibitors and low concentrations of actinomycin D on ribosomal RNA synthesis. FEBS Lett. 1979;107:281–284. doi: 10.1016/0014-5793(79)80390-7. [DOI] [PubMed] [Google Scholar]
- 14.Albanèse V., Reissmann S., Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 2010;189:69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greber B.J., Boehringer D., Montellese C., Ban N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2012;19:1228–1233. doi: 10.1038/nsmb.2425. [DOI] [PubMed] [Google Scholar]
- 16.Meyer A.E., Hung N.J., Yang P., Johnson A.W., Craig E.A. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demoinet E., Jacquier A., Lutfalla G., Fromont-Racine M. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA. 2007;13:1570–1581. doi: 10.1261/rna.585007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahi C., Craig E.A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer A.E., Hoover L.A., Craig E.A. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J. Biol. Chem. 2010;285:961–968. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squatrito M., Mancino M., Donzelli M., Areces L.B., Draetta G.F. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 21.Pilipenko E.V., Pestova T.V., Kolupaeva V.G., Khitrina E.V., Poperechnaya A.N., Agol V.I., Hellen C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 22.Xia X., Cheng A., Lessor T., Zhang Y., Hamburger A.W. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell. Physiol. 2001;187:209–217. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- 23.Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 24.Squatrito M., Mancino M., Sala L., Draetta G.F. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem. Biophys. Res. Commun. 2006;344:859–868. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 25.Narla A., Ebert B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulima S.O., Patchett S., Advani V.M., De Keersmaecker K., Johnson A.W., Dinman J.D. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc. Natl. Acad. Sci. USA. 2014;111:5640–5645. doi: 10.1073/pnas.1400247111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Keersmaecker K., Sulima S.O., Dinman J.D. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood. 2015;125:1377–1382. doi: 10.1182/blood-2014-10-569616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson E., Ferreira-Cerca S., Hurt E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013;126:4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 29.Menne T.F., Goyenechea B., Sánchez-Puig N., Wong C.C., Tonkin L.M., Ancliff P.J., Brost R.L., Costanzo M., Boone C., Warren A.J. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 30.Weis F., Giudice E., Churcher M., Jin L., Hilcenko C., Wong C.C., Traynor D., Kay R.R., Warren A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong C.C., Traynor D., Basse N., Kay R.R., Warren A.J. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118:4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 32.Lo K.Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shammas C., Menne T.F., Hilcenko C., Michell S.R., Goyenechea B., Boocock G.R., Durie P.R., Rommens J.M., Warren A.J. Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond Syndrome. J. Biol. Chem. 2005;280:19221–19229. doi: 10.1074/jbc.M414656200. [DOI] [PubMed] [Google Scholar]
- 34.Dror Y., Freedman M.H. Shwachman-diamond syndrome. Br. J. Haematol. 2002;118:701–713. doi: 10.1046/j.1365-2141.2002.03585.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S4 and Tables S1–S3
Document S2. Article plus Supplemental Data