Initial seeding of embryonic thymus by immune-restricted lympho-myeloid progenitors (original) (raw)
. Author manuscript; available in PMC: 2017 Jun 1.
Published in final edited form as: Nat Immunol. 2016 Oct 3;17(12):1424–1435. doi: 10.1038/ni.3576
Abstract
The last stages of T-lineage-restriction occur in the thymus following entry of thymus-seeding progenitors (TSPs). The identity and lineage potentials of TSPs remains unclear. Since the first embryonic TSPs enter a non-vascularized thymus-rudiment, we were able to directly image and establish the functional and molecular properties of embryonic thymopoiesis-initiating progenitors (T-IPs) prior to thymus-entry and Notch-activation. T-IPs did not include multipotent stem cells nor molecular evidence of T-cell-restricted progenitors. Rather, single cell molecular and functional analysis demonstrated that most fetal T-IPs co-express programs and potentials for lymphoid and myeloid components of the immune-system. Moreover, studies of _Rbpj_-deficient embryos, demonstrate, contrary to previous claims, that canonical Notch-signaling is neither involved in pre-thymic T-lineage-restriction nor T-IP migration.
Multiple distinct T lymphocyte subsets which function as critical mediators of the adaptive immune-system, all derive from multipotent hematopoietic stem cells (HSCs). Dysregulated T lymphocyte development underlies severe immune-deficiencies as well as T cell leukemias.
In mammalian hematopoiesis, the final steps towards T-lymphocyte lineage restriction take place in the thymus1 unlike other blood cell lineages generated from HSCs, which occurs within the bone marrow (BM). Thymopoiesis is therefore normally maintained through replenishment by thymus-seeding progenitors (TSPs) migrating from the BM to the thymus1. Establishing the identity and functional and molecular properties of TSPs is therefore critical towards understanding which T-lineage-restriction steps, from fully multipotent HSCs to T-cell restricted progenitors, occur in the BM and subsequently in the distant thymus. Identification and molecular characterization of TSPs are also key towards further unraveling the molecular cues guiding this critical lineage-restriction process as well as the pathways promoting TSP migration and transition from the BM to the thymus.
Despite of extensive investigations, the identity and lineage potentials of TSPs remain disputed, largely reflecting that BM-derived TSPs migrate through the circulation to the vascularized thymus. Therefore it has not been possible to directly study or image BM-derived mammalian TSPs prior to thymus entry. Rather, extensive efforts aimed at obtaining a better understanding of the identity and regulation of TSPs, focused on characterization of early thymic progenitors (ETPs), already residing within the thymus1, 2, 3, 4, therefore only allowing inferences about TSPs1, 5. This caveat is particularly relevant for studies that failed to identify HSCs in the postnatal or adult thymus, interpreted to imply that TSPs do not include HSCs3, 6. However, since thymic epithelium expresses high levels of Notch ligands7, which can rapidly induce T-lineage-restriction of multipotent stem/progenitor cells7, in the absence of direct evidence, the possibility that HSCs might act as TSPs cannot be excluded.
In the mouse the thymic rudiment develops from around embryonic day (E) 98 and is first seeded by hematopoietic cells around E11.59. In contrast to later fetal development and postnatally, when TSPs reach the thymus through the circulation5, at E11.5 the BM is still not established and the thymus rudiment is not yet vascularized. Therefore, the first embryonic TSPs are recruited to the thymus rudiment by migration through the surrounding mesenchyme5, guided by chemokines produced in the thymic anlage10. Canonical Notch-signaling is critically involved in the earliest stages of T-cell lineage restriction in fetal and adult thymopoiesis11, 12, 13, 14, and has also been suggested to promote migration of TSPs from the BM to the thymus15, although no studies have directly investigated the role of Notch-signaling in the initial seeding of the embryonic thymic rudiment. Importantly, previous studies concluded that TSPs might already be Notch-activated in the fetal liver (FL), prior to migration to the embryonic thymus, and that Notch-signaling could mediate pre-thymic T-lineage restriction of FL progenitors16, 17, although this has yet to be investigated in embryos deficient in canonical notch signaling.
The lineage potentials of the first thymopoiesis-initiating progenitors (T-IPs), responsible for initiation of embryonic thymopoiesis remain unclear, although the thymus harbors multiple non-T- cell lineages proposed to play a role in supporting T-cell development18, including removal of apoptotic thymocytes by monocytes/macrophages19. In the adult thymus, fate mapping studies suggested that monocytes mostly develop independently of ETPs2, 20, 21, whereas the origin of monocytes/macrophages residing in the embryonic thymus22 remains to be investigated.
While some studies suggested that HSCs might be responsible for initial seeding of the embryonic thymus23, later studies failed to support this24, albeit investigating the thymus at E12 when Notch-ligands are already expressed16. Other studies have suggested that embryonic TSPs might lack myeloid and B-lymphocyte potential16, 24, 25 and that embryonic TSPs, unlike adult TSPs1, 3, might be largely T-cell-restricted prior to seeding the thymus. Regardless, no previous studies have at the single cell level, molecularly and functionally characterized the first mammalian T-IPs, as they migrate through the mesenchyme towards the thymic rudiment.
Herein, we establish that the first T-IPs migrating through the mesenchyme surrounding the thymus anlage, do not include HSCs, but express the lymphoid reporter _Rag1_-GFP3, 26, already prior to thymus-entry. Most of these T-IPs, despite their uniform lymphoid gene expression, are not yet lymphoid-lineage restricted, as they also express myeloid lineage programs and potential, and contribute to myeloid as well as T-cell development in the embryonic thymus. Moreover, their initial colonization of the thymus rudiment occurs independently of canonical Notch signaling. Our findings establish that embryonic thymopoiesis is initiated by lympho-myeloid restricted progenitors.
Results
The embryonic thymus rudiment is seeded for the first time at E11.25
Based primarily on timed matings, colonization of the mouse thymus rudiment has been reported to occur around E11-11.59, 25. However, the developmental stage based on timed matings between and within different litters, and between strains, can vary significantly27. Moreover, from E11 the somite pairs are not clearly visible. To more accurately define the developmental stage at the time of initial thymic seeding, we established a more detailed staging of E11.0-11.75 embryos based on the number of tail somites (TS), using the cloaca as a landmark (Fig. 1a and Supplementary Fig. 1a,b). This allowed staging of embryos at 0.25 day increments. In the E11.5 mouse embryo, the common thymus and parathyroid rudiments are located bilaterally in the third pharyngeal arches, attached to the endoderm and surface ectoderm28, and adjacent to the aortic arches marked by VE-Cadherin and the endothelial reporter Von Willebrand factor (Vwf)-eGFP (Supplementary Fig. 1c-e). Following tail somite staging, dissected thymic lobes were carefully cleaned from surrounding blood vessels to avoid blood contamination (Fig. 1b and Supplementary Fig. 1e and 2a). Subsequently, colonization of the thymic rudiment by the first hematopoietic cells with T-lymphocyte potential was consistently observed at TS10-12 (corresponding to E11.25-E11.5; Fig. 1a), when approximately 50% of thymic rudiments contained T-cell potential (Fig. 1c,d).
Figure 1. Initial seeding of the embryonic thymic anlage.
(a) Correlation of the number of tail somites with the timing of embryonic development. Each circle represents an individual embryo. Area between broken lines corresponds to the tail somite stages (10-12) of initial thymus seeding.
(b) Typical dissected thymic lobes from E11.5 embryos stained with VE-Cadherin (VE-Cad, red), cytokeratin (CK, green) and DAPI (blue). VE-Cadherin+ endothelial cells were completely removed by dissection in most explants (top), but not all (bottom).
(c) Frequency of seeded embryonic thymic rudiments, as determined by emergence of T cell potential of dissected thymuses at tail somites 4-9 (_n_=18), 10-12 (_n_=17) and 13-16 (_n_=9), corresponding to E11.0-E11.75 embryos. p values were calculated with two-sided Fisher’s exact test.
(d) Representative FACS profile of T cells (CD4+CD8+ and/or Thy1.2+CD25+) derived from culture of an individual E11.5 (TS11) thymus on OP9-DL1 stroma.
(e) Expression (2–ΔCt) of Dll4 in PLET1+ thymic epithelial cells isolated from E11.5 and E12.5 embryos. Mean (s.d.) expression levels (relative to the average of Actb, Hmbs and Tbp) of 2 biological replicates are shown. **p<0.01.
As Notch activation might restrict lineage potentials of T-IPs, we investigated embryonic PLET1+ thymic epithelial cells (TECs)8 for expression of delta-like 4 (Dll4), encoding the Notch ligand critical for embryonic and adult T-cell development11, 29. Whereas in agreement with previous studies16 Dll4 was expressed in E12.5 PLET1+ TECs, it was much lower at E11.5 (TS11-14) (Fig. 1e), consistent with Foxn1 expression being essential for induction of Dll4 expression30 after E11-E11.58.
Multipotent HSCs do not initiate embryonic thymopoiesis
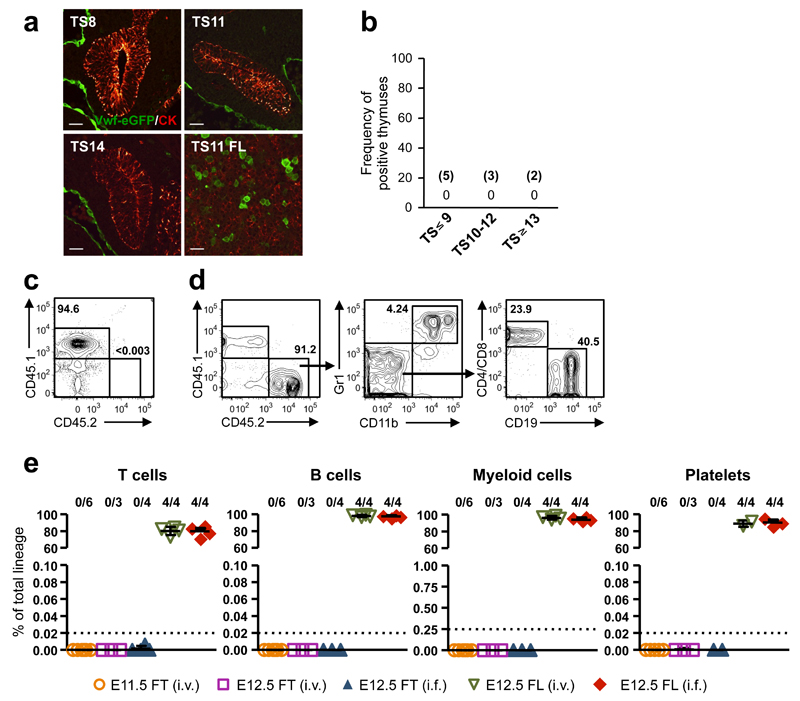
At E11.5, T-IPs migrate through the surrounding mesenchyme5 to colonize the thymic rudiment, allowing imaging of candidate T-IPs prior to thymus-entry and preceding Notch-activation. There has been considerable disagreement about the identity and lineage potentials of progenitors responsible for the initial seeding of the embryonic thymus, ranging from multipotent stem/progenitor cells to T-cell restricted progenitors16, 23, 25. Since all definitive HSCs in the FL express Vwf31, in contrast to fetal lympho-myeloid-restricted progenitors32, we used a _Vwf_-eGFP reporter mouse31 to investigate whether HSCs migrate towards the thymic rudiment in connection with its initial seeding at TS10-12. _Vwf_-eGFP expression was confirmed by co-staining with an anti-Vwf antibody (Supplementary Fig. 2b). Despite complete sectioning of multiple thymuses before, during and immediately after their initial seeding by T-IPs, we found no evidence of any _Vwf_-eGFP+ cells migrating towards the thymus primordium (Fig. 2a,b). Consistent with this, both E11.5 and E12.5 thymocytes failed to long-term reconstitute irradiated mice in competitive (Fig. 2c-e) as well as non-competitive (Supplementary Fig. 2c-f) transplantations, unlike E12.5 FL cells which contained robust HSC-activity (Fig. 2c-e and Supplementary Fig. 2c-f). These results corroborate the absence of definitive HSCs among T-IPs colonizing the thymus primordium at TS10-12.
Figure 2. Hematopoietic stem cells do not seed the embryonic thymus rudiment.
(a) Vwf eGFP (green; marking HSCs and endothelium) and cytokeratin (CK, red) staining in thymus and FL sections of TS8-14 Vwf eGFP+ embryos. Scale bars: 10µm.
(b) Frequency of E11.0-E11.75 embryos with Vwf eGFP_+_ inside and/or adjacent to/lining the thymus rudiment. The number of embryos investigated is shown in brackets. In each case the complete thymuses were sectioned and analyzed.
(c-e) Long-term (HSC) reconstituting activity of total thymocytes (fetal thymus; FT) from 4-5 E11.5 or E12.5 Vwf eGFP embryos or total FL (FL) cells from 3 E12.5 Vwf eGFP embryos, transplanted intravenously (i.v.) or intrafemorally (i.f.) into each lethally irradiated recipient. (c,d) FACS profile of typical peripheral blood of a CD45.1 mouse transplanted i.f. with CD45.2+ E12.5 total fetal thymocytes (c) or with CD45.2+ E12.5 total FL cells (d) 16 weeks earlier. No thymocyte-derived (CD45.2+) cells were observed (detection level 0.003%). Left, percentage CD45.2 contribution to total blood cells, Middle and right, distribution between myeloid cells (Gr1+CD11b+), B cells (CD19+) and T cells (CD4/CD8+), within CD45.2+ cells. (e) Summary of long-term thymocyte and FL reconstitution of blood cell lineages, 15-17 weeks post-transplantation, as percentage of total cells within each lineage. Numbers above graphs indicate frequency of reconstituted mice (see Online Methods). Dotted lines indicate the detection level of reconstitution for each lineage, based on the number of events acquired by FACS. Data from 5 independent experiments.
Initial seeding of the embryonic thymus rudiment by _Rag1_-expressing progenitors
To investigate whether partially or fully lymphoid-restricted progenitors migrate towards the thymic rudiment at or before TS10-12, we used a _Rag1-_driven GFP reporter mouse26. _Rag1-_GFP+ cells first appeared in the thymus rudiment at TS10-12 (Fig. 3a,b), coinciding with the first colonization of the thymic rudiment by hematopoietic cells harboring T-cell potential (Fig. 1c,d). Notably, _Rag1_-GFP+ cells first appeared in low numbers in the mesenchyme immediately outside (lining) the epithelial thymic rudiment, and shortly thereafter inside the thymus primordium (Fig. 3a,b and Supplementary Fig. 2g-i). Further characterization of E11.5 (TS10-14) _Rag1_-GFP+ cells revealed uniform expression of CD45, c-Kit, Flt3 and IL-7Rα, and absence of CD25 (Fig. 3c,d), similar to multipotent ETPs in the postnatal thymus3, and recently identified lympho-myeloid-restricted progenitors in E10.5-11.5 FL, which in addition to B and T-lymphocyte potential also sustain granulocyte-macrophage (GM) but not megakaryocyte-erythroid (MkE) potential, and therefore are entirely immune-restricted in their lineage fate options32. _Rag1_-GFP+ cells also expressed the chemokine (C-C motif) receptor (CCR) 9 (Fig. 3c), critical for embryonic thymus colonization10. Virtually all cells expressing the pan-hematopoietic stem/progenitor receptor c-kit in the E11.5 (TS10-14) thymic rudiment, were _Rag1-_GFP+ (not shown), suggesting that initial seeding of the thymic rudiment is largely restricted to CD45+Lin−c-Kit+CD25−Flt3+CCR9+_Rag1_-GFP+progenitors. To investigate whether these _Rag1_-GFP+ progenitors are able to progress through the normal thymic progenitor stages emerging in the E13.5-E14.5 thymus26, we cultured whole thymic lobes from E11.5 _Rag1_-GFP+ embryos, and obtained evidence that _Rag1_-GFP+ progenitors can differentiate into all double negative (DN) thymocyte stages (DN1-4) and double positive (DP) CD4+CD8+ T cell progenitors (Fig. 3e).
Figure 3. Initial seeding of the embryonic thymus by _Rag1-_GFP+ progenitors.
(a) Representative _Rag1-_GFP (green) expression in cytokeratin (CK, blue) stained sections of the thymus rudiment in TS8-16 _Rag1_-GFP+ embryos. Scale bars represent 10µm. (also see Supplementary Fig. 2g-i).
(b) Frequencies of E11.0-E11.75 embryos with Rag1-GFP+ cells exclusively lining (maximum 2 cell diameter distance from thymic epithelium) or lining and inside the thymus rudiment. Embryos with _Rag1_-GFP+ cells inside the thymic rudiment consistently also showed _Rag1_-GFP+ cells outside or lining the thymus. The number of individual embryos investigated shown in brackets. *; 3 complete thymuses were serially sectioned and analyzed, whereas remaining thymuses were partially analyzed (see Online Methods).
(c,d) Representative FACS profiles of pooled E11.5 thymuses. Virtually all _Rag1_-GFP+ cells express CD45 and progenitor markers c-Kit, Flt3 and IL-7Rα but are negative for lineage markers and CD25. The vast majority is also CCR9+. The number in the first plot in c, reflects the percentage of total cells, whereas subsequent plots show frequencies relative to total GFP+ cells. In d, numbers reflect frequencies relative to CD45+_Rag1_-GFP+ cells. All frequencies are means of 2 independent experiments, 8 and 9 embryos pooled in each experiment.
(e) Analysis of T-cell progenitor development from E11.5 T-IPs on OP9-DL1 stroma. Representative FACS profiles after 7 and 14 days of culture, of cells generated from individual E11.5 thymic lobes isolated at indicated developmental stages. Data correspond to 3 diffferent embryos, representative of a total of 9 analyzed embryos. Numbers in plots indicate percentages of total CD45+ cells.
_Rag_-_1_-expressing T-IPs have immune-restricted but combined lympho-myeloid potential
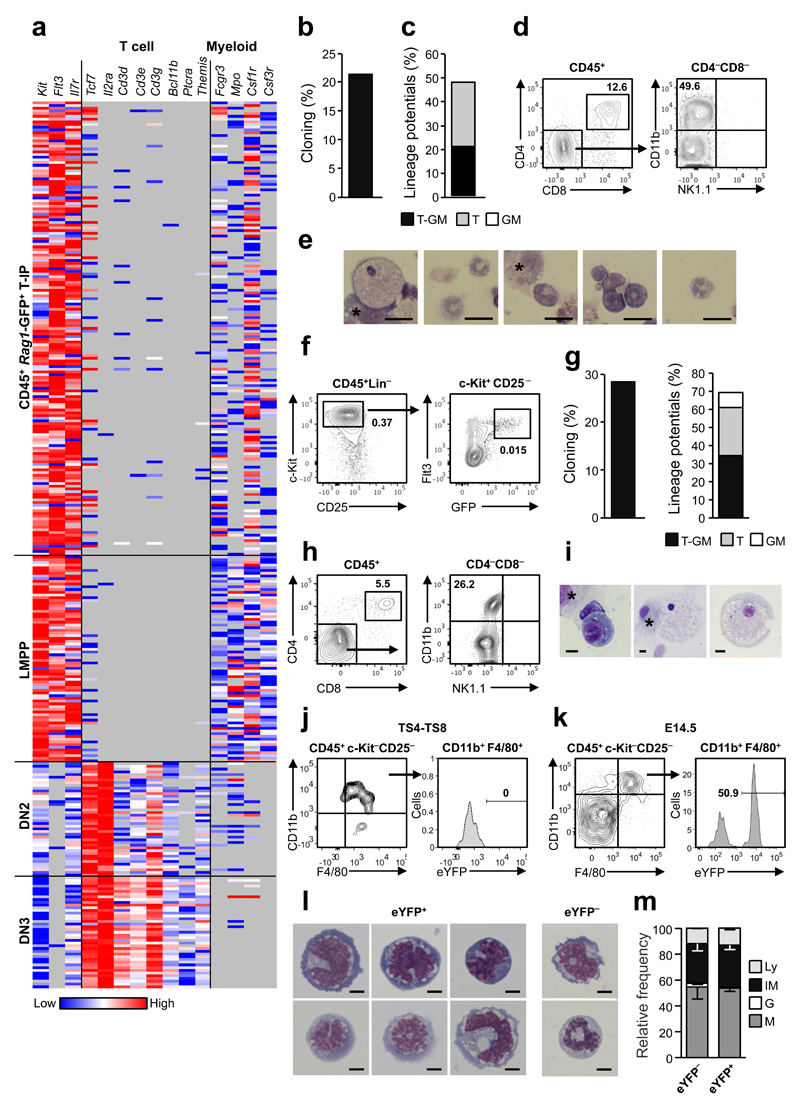
To investigate whether _Rag1_-GFP+ cells seeding the E11.5 thymus rudiment are multipotent or T-cell-restricted, we next performed single cell gene expression analysis. The expression of distinct T-cell and myeloid genes in CD45+_Rag1_-GFP+ T-IPs was compared to that of lymphoid-primed multipotent progenitors (LMPPs) from E11.5 FL32, as well as E14.5 partially T-cell-restricted DN2 and fully T cell-restricted DN3 thymic progenitors (Fig. 4a and Supplementary Fig. 2j,k). Expression profiles of E11.5 T-IPs and LMPPs were strikingly similar, with virtually no detectable expression of T-cell-restricted genes, but with co-expression of early lymphoid and myeloid genes, in contrast to DN3 T-cell-restricted rogenitors1. These expression signatures are most compatible with the thymus anlage being seeded primarily by _Rag1-_expressing T-IPs with sustained myeloid lineage potential. In agreement, at E12.5 when CD45+Lin−c-Kit+CD25−Flt3+ ETPs are more frequent, they revealed high T-cell and myeloid lineage potential in vitro (Supplementary Fig. 3a-c). Similarly, single cell cultures of E11.5 CD45+Lin–c-Kit+CD25−Flt3+ T-IPs demonstrated combined T and myeloid lineage potential (Fig. 4b-e and Supplementary Fig. 3d-f). Progenitors with the same CD45+Lin−c-Kit+CD25−Flt3+ _Rag1_-GFP+ phenotype were identified in the blood circulation of E11.5 embryos and had comparable frequencies of combined T and myeloid lineage potential as T-IPs (Fig. 4f-i and Supplementary Fig. 3g,h).
Figure 4. Thymopoiesis-initiating progenitors have combined T-lymphoid and granulocyte-monocyte lineage potentials.
(a) Cytokine receptor, T cell and myeloid lineage affiliated gene expression (ΔCt, relative to Hprt) in single E11.5 CD45+_Rag1_-GFP+ T-IPs (_n_=166), E11.5 CD45+Lin−B220−CD19−c-Kit+Flt3+IL-7Rα+ FL lympho-myeloid-restricted progenitors (_n=_77), E14.5 Lin−CD4−CD8−c-Kit+CD25+ DN2 (_n_=42) and E14.5 Lin−CD4−CD8−c-Kit−CD25+ DN3 (_n_=41), purified from 2-4 biological replicates, each representing 8-16 embryos. Grey, below detection level. Only Hprt amplified cells (98.8%) were included.
(b,c) Cloning frequency (b) and lineage distribution (c) of single CD45+Lin−c-Kit+CD25−Flt3+ E11.5 T-IPs cultured on OP9-DL1 (_n_=88 cells), from 4 experiments.
(d,e) Representative clone from single CD45+Lin−c-Kit+CD25–Flt3+ E11.5 T-IP on OP9-DL1 producing CD4+CD8+ T cells (d) and myeloid cells confirmed by FACS (CD11b+CD4−CD8−NK1.1−; d) and morphology (e). Asterisk in e, indicate OP9-DL1. Scale bars: 20µm.
(f) E11.5 circulating CD45+Lin−c-Kit+CD25−Flt3+_Rag1_-GFP+ cells. Mean percentages of total live cells from 4 litters, 7-9 embryos each.
(g) Frequency of clones (left) and myeloid and/or T-cell potential (right), from single circulating E11.5 CD45+Lin−c-Kit+CD25−Flt3+_Rag1_-GFP+ cells on OP9-DL1. Means of 3 independent experiments (_n_=254 cells).
(h,i) Representative clone from single circulating E11.5 CD45+Lin−c-Kit+CD25−Flt3+_Rag1_-GFP+ cell producing CD4+CD8+ T-cells (h) and myeloid cells (h-i). * = OP9-DL1 cells. Scale bars: 10µm.
(j,k) _Rag1_-Cre fate mapping of myeloid cells from pooled TS4-TS8 (j, _n_=3 experiments) and E14.5 (k,_n_=2 experiments) _Rag1_-Cretg/+_R26_eYFP/+ thymuses, showing eYFP expression (mean frequency) on CD11b+F4/80+c-Kit−CD25− myeloid cells in E14.5 but not TS4-TS8 thymuses.
(l) Morphology of representative eYFP+ and eYFP− CD11b+F4/80+c-Kit−CD25− monocytic cells from E14.5 _Rag1_-Cretg/+_R26_eYFP/+ thymuses. Scale bars: 5µm.
(m) Frequencies of monocytes/macrophages (M), granulocytes (G), Immature myeloid (IM) and lymphocytes (Ly) determined by morphology of sorted E14.5 eYFP− or eYFP+ CD11b+F4/80+c-Kit− CD25− cells. Mean (s.d.) results from 2 experiments (4-6 pooled embryos; _n=_152 eYFP− cells and _n=_199 eYFP+ cells).
Recent studies suggested that ETPs might be an important source of granulocytes but not monocytes in the adult thymus2, 20. To investigate whether _Rag1-_GFP+ T-IPs contribute physiologically to myeloid cells in the early embryonic thymus, we performed _Rag1-_Cre fate-mapping33, 34. Analysis of thymus rudiments before thymus colonization (TS4-8), revealed that CD11b+F4/80+ monocytes/macrophages in the thymic rudiment prior to its seeding by T-IPs22, were not derived from _Rag1_-expressing cells (Fig. 4j). In contrast, analysis of E14.5 embryos suggested that as much as 50% of CD11b+F4/80+ monocytes/macrophages in the thymus were derived from _Rag1-_expressing progenitors (Fig. 4k-m and Supplementary Fig. 3i). In contrast, analysis of E14.5 _Rag1_-GFP thymuses, showed strong GFP-expression in all ETPs, DN2 and DN3 thymocyte progenitors, but no expression in CD11b+F4/80+ macrophages, supporting that the eYFP expression in macrophages in _Rag1_-CreTg/+_R26_-stop-eYFPFl/+ E14.5 thymuses is not derived from eYFP expressed by potentially phagocytosed thymocytes (Supplementary Fig. 3j-l). Following transplantation of lethally irradiated CD45.1 mice with a mixture of _Rag1_-CreTg/+ and _R26_-stop-eYFPFl/Fl CD45.2 BM cells, thymic progenitors and monocytes/macrophages were almost entirely donor-derived (Supplementary Fig. 3m). The absence of eYFP-expression in these thymic monocytes/macrophages (Supplementary Fig. 3n) suggests that potentially phagocytosed _Rag1_-Cre-expressing thymocytes do not result in recombination in floxed _R26_eYFP macrophages. These results demonstrate that following embryonic thymus colonization, a large proportion of thymic monocytes/macrophages are derived from _Rag1_-expressing progenitors.
In contrast to the high T-cell and granulocyte-monocyte (GM) lineage potentials, Mk and E potential was virtually absent in E11.25-E11.75 thymuses (Fig. 5a and Supplementary Fig. 4a), despite abundant MkE-potential in the FL (Supplementary Fig. 4b,c). The lack of significant MkE-potential also confirmed that meticulous dissection had effectively avoided progenitor contamination from blood vessels surrounding the thymus (Fig. 1b and Supplementary Fig. 2a). Specifically, in a total of 18 thymuses analyzed for Mk-potential, only two small cell clusters positive for acetylcholinesterase staining were identified, and from 17 thymuses not a single E-colony was found.
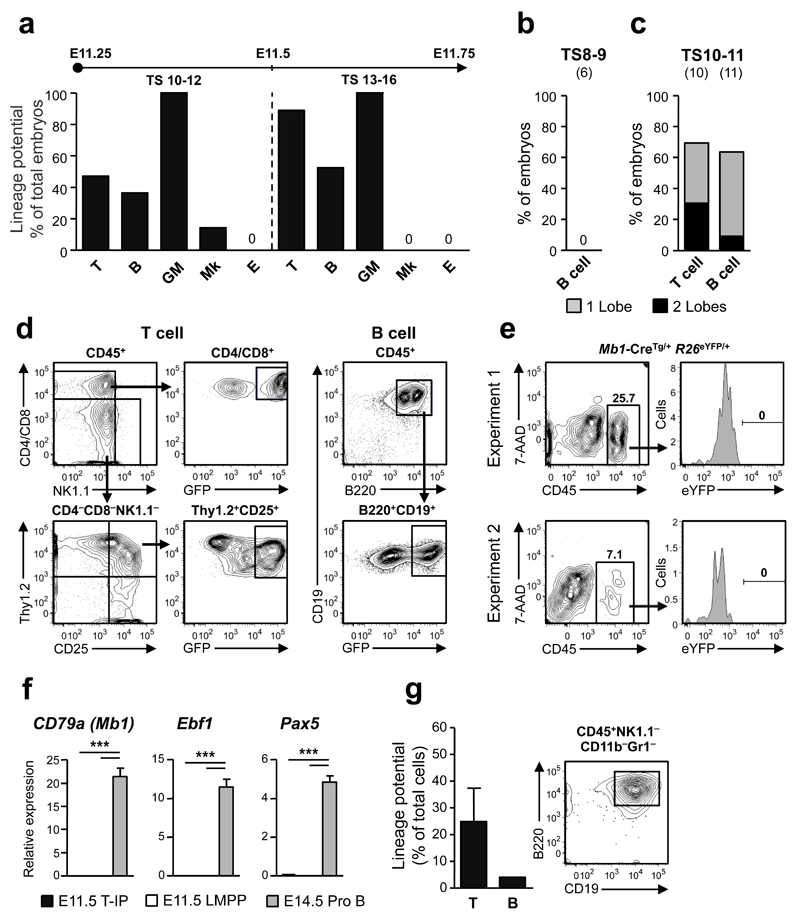
Figure 5. Embryonic thymopoiesis is initiated by _Rag1-_GFP+ lympho-myeloid restricted progenitors.
(a) T cell, B cell, myeloid (granulocyte-macrophage; GM), megakaryocyte (Mk) and erythroid (E) potential of dissected individual thymuses from embryos at TS10-12 (_n_=7-22 embryos investigated for each lineage) and TS13-16 (_n_=9-21 embryos).
(b-d) B cell (TS8-9 and TS10-11) and T cell (TS10-11) potential of individual thymic lobes following culture on OP9 and OP9-DL co-cultures, respectively. Shown is the percentage of embryos with one (grey) or both (black) thymic lobes exhibiting T cell (CD4/CD8+_Rag1_-GFP+ and/or Thy1.2+CD25+_Rag1_-GFP+) and B cell (B220+CD19+_Rag1_-GFP+) potential, respectively. The number of embryos investigated is shown in brackets. (d) Typical FACS profiles of cultures with individual thymic lobes showing T cell and B cell potential (see Supplementary Fig. 4d,e for additional profiles).
(e) Analysis of pooled E11.5 _Mb1_-Cretg/+_R26_eYFP/+ embryos showing lack of eYFP expression in CD45+ thymocytes (_n=_3 and n=5 eYFP+ embryos per pool in 2 independent experiments).
(f) Single cell gene expression of early B-cell specific genes in E11.5 T-IPs (_n_=166 cells), E11.5 fetal liver CD45+Lin–B220–CD19–c-Kit+Flt3+IL-7Rα+ LMPPs (_n_=77 cells) and E14.5 Lin− B220+CD43+CD19+CD24+AA4.1+IL-7Rα+c-Kit+ ProB-cells (_n_=41 cells) from 2 biological replicates, shown as mean (s.d.) expression values (relative to Hprt). ***p<0.001
(g) Left; Mean (s.e.m.) frequencies of FACS purified single CD45+Lin−c-Kit+CD25−Flt3+ E12.5 thymic progenitors from VavP-_Mcl1_–transgenic mice, possessing T cell and B cell potential (_n_=96 single cells from 2 experiments). Right; B220+CD19+ B cells generated from a single E12.5 CD45+Lin–c-Kit+CD25– Flt3+ cell on OP9 stroma cells.
Contrary to previous studies inferring that embryonic ETPs lack B-cell potential16, 17, 24, 25, 35, distinguishing them from multipotent postnatal ETPs3, 17, we found evidence for B-cell potential emerging in the thymus at the exact time of its colonization by T-IPs (TS10-12) (Fig. 5a-d). Importantly, since B-cell-restricted progenitors are yet to emerge anywhere in the E11.5 embryo36, any uncovered B-cell potential likely derives from T-IPs which also have T-lineage potential. Since processing of the embryonic thymus rudiment followed by FACS-based cell sorting might compromise the ability of T-IPs to read out in lineage potential assays, we first investigated T and B-cell potential in whole dissected individual TS10-11 thymic lobes (Fig. 1b), when approximately 50% of embryos have detectable T-cell potential (Fig. 1c). No B-cell potential was identified in TS8-9 thymic rudiments (Fig. 5b), whereas at TS10-11, the frequency of thymuses with T and B-cell potential was comparable (70% and 64%, respectively; Fig. 5c,d and Supplementary Fig 4d,e), corroborating that at least a fraction of T-IPs seeding the thymus rudiment have combined T and B-lineage potential. The absence of B-cell-restricted progenitors in the E11.5 thymic rudiment was further supported by fate-mapping with Mb1_-Cre,_ active from the earliest stage of B-cell-restricted progenitors37 (Fig. 5e). Moreover like E11.5 FL LMPPs, E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs lacked expression of Mb1, Ebf1 and Pax5 (Fig. 5f). Intriguingly, we failed to detect significant B-cell potential from FACS-purified wildtype (WT) CD45+Lin−c-Kit+CD25−Flt3+ cells (Luc et al, unpublished data). However, since this potential was clearly present in cultures of whole thymus rudiments (Fig 5a-d), we next used mice expressing Mcl1 to enhance cell survival3, and doing so detected definitive B-cell potential from a low fraction of purified single CD45+Lin−c-Kit+CD25−Flt3+ cells (Fig. 5g), supporting that some T-IPs also possess B cell potential.
Molecular profiling of E11.5 thymopoiesis-initiating progenitors
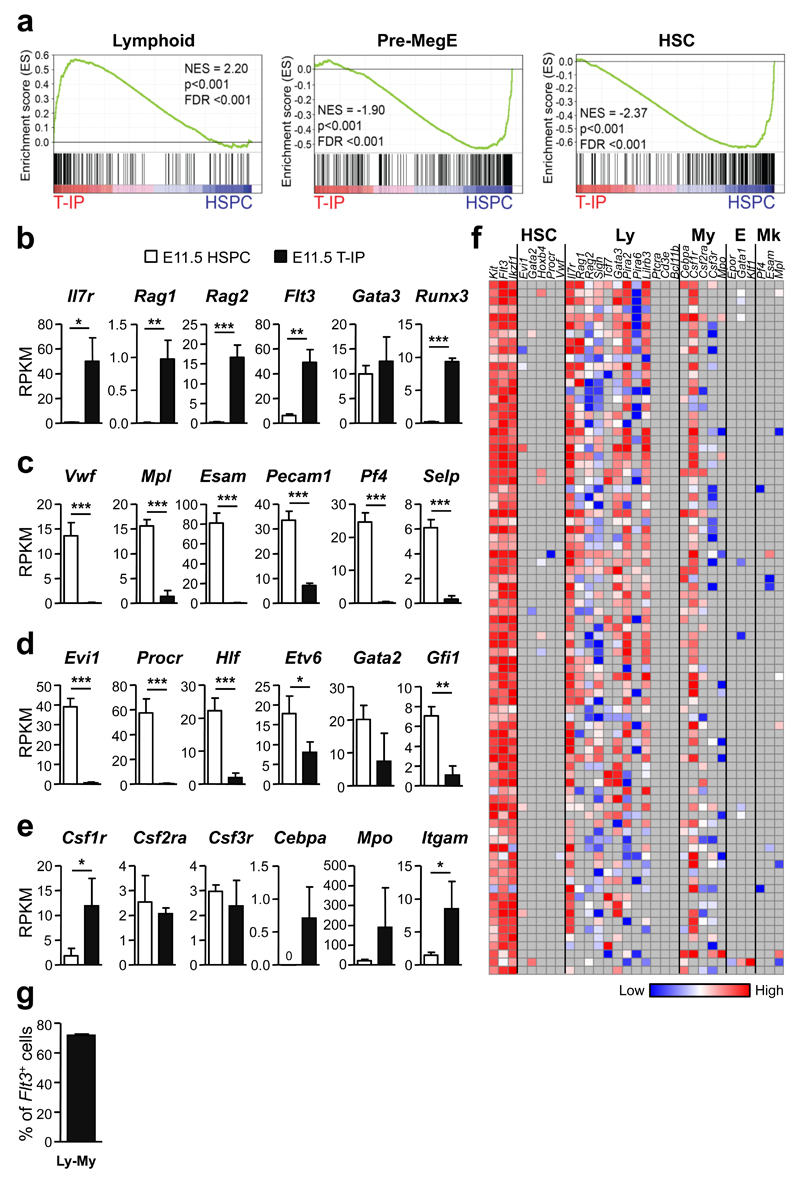
Gene-set enrichment analyses (GSEA) using published gene sets3, 38 of RNA sequencing data32 from E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs and E11.5 Lin−CD45loVE-Cad+c-Kit+ hematopoietic stem/progenitor cells (HSPCs)39 from the aorta-gonad-mesonephros (AGM) region (Supplementary Fig. 4f) demonstrated highly significant up-regulation of typical early lymphoid genes, and down-regulation of MkE and HSC genes in E11.5 T-IPs (Fig. 6a-d). Many myeloid genes were also distinctly upregulated in E11.5 T-IPs (Fig. 6e). Significantly upregulated genes in E11.5 T-IPs compared to E11.5 HSPCs were notably over-represented in immune-related processes (Supplementary Tables 1 and 2), including pathways involved in chemotaxis.
Figure 6. Molecular profiling of E11.5 thymopoiesis-initiating progenitors.
(a) Gene-set enrichment analyses of global RNA sequencing data from E11.5 CD45+Lin−c-Kit+CD25− Flt3+ T-IPs (_n_=3) versus E11.5 Lin−CD45loVE-Cad+c-Kit+ AGM stem/progenitor cells (HSPC) (_n_=3) for Lymphoid, Pre-MegE and HSC gene sets. NES, normalized enrichment score; FDR, false discovery rate.
(b-e) mRNA expression, shown as mean (s.d.) RPKM, of (b) early lymphoid (c) Mk (d) HSC and (e) GM affiliated genes in E11.5 T-IPs (_n_=3) and AGM HSPCs (_n_=3). *p<0.05, **p<0.01, ***p<0.001. n represents number of biological replicates.
(f) Heatmap for expression (ΔCt values, relative to Hprt) of lineage (HSC, hematopoietic stem cell; Ly, lymphoid; My, myeloid; E, erythroid; Mk, megakaryocytic) affiliated genes in single E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs (_n_=85 cells, from 2 biological replicates, each using a pool of 5 and 9 embryos). Grey indicates not detected. Three cells were excluded due to absence of Flt3 amplification.
(g) Mean (s.d.) frequency of single E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs co-expressing GM and lymphoid affiliated genes while not expressing Mk or E genes (from f). Only cells that amplified Flt3, c-Kit and Hprt (corresponding to 97% of total cells analyzed) were included. Flt3 and Gata3 were not considered as lymphoid in this analysis due to their concomitant non-lymphoid expression pattern.
To seek further molecular support for combined myeloid and lymphoid lineage potential in T-IPs, we performed single cell gene expression analysis of E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs. All cells showed expression of multiple lymphoid genes, and 70% of single E11.5 T-IPs co-expressed lymphoid and myeloid genes, but not MkE genes (Fig. 6f,g). Moreover, genes defining the earliest T-cell-restricted progenitors3, 40, including Ptcra, Cd3e and Bcl11b were not expressed in E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs (Fig. 6f; see also Fig. 4a). These molecular findings corroborate that E11.5 T-IPs have immune-restricted but typically combined lympho-myeloid lineage potentials when seeding the embryonic thymic rudiment. Furthermore, these molecular data, in particular the lack of expression of genes uniformly expressed in the earliest T-cell restricted thymic progenitors, are most compatible with few if any T-IPs being fully T-cell-restricted prior to thymus entry.
Ontogeny-related changes in thymus-seeding progenitor pathways
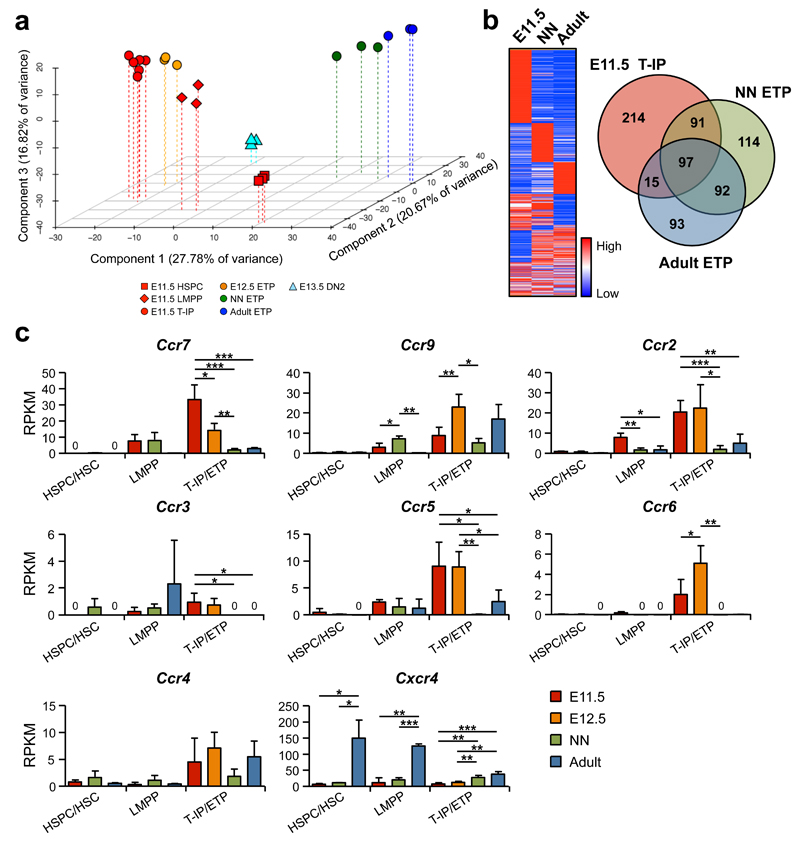
Principal component analysis (PCA) and hierarchical clustering analysis demonstrated that E11.5 T-IPs clustered more closely to (and between) the recently described E11.5 FL LMPPs32 and E12.5 ETPs (Supplementary Fig. 4g) than to neonatal (1 week) and adult (8 weeks) ETPs, and more distant to the more multipotent E11.5 AGM HSPCs as well as the first E13.5 DN2s (Fig. 7a and Supplementary Fig. 5a,b). This supports that T-IPs seeding the thymic anlage might be derived from FL lympho-myeloid restricted progenitors and give rise to the first intra-thymic ETPs. When comparing E11.5 T-IPs with neonatal and adult ETPs, and the corresponding HSPC/HSC populations, 97 genes were highly up-regulated in the T-IP/ETP populations, irrespectively of developmental stage (Fig. 7b). These included Ccr7 and Ccr9, previously shown to be involved in the migration of fetal and adult TSPs to the thymus10, 41 (Fig. 7b,c and Supplementary Table 3). In addition we identified genes uniquely upregulated in E11.5 T-IPs compared to HSPCs/HSCs, neonatal and adult ETPs (Fig. 7b and Supplementary Tables 4-6). Interestingly, these included additional chemokine receptor genes, implicating usage of distinct chemokine receptors by embryonic T-IPs. Specifically, in addition to Ccr7, expression of Ccr2, Ccr5 and Ccr6 was notably higher in E11.5 T-IPs and in E12.ETPs, than neonatal and adult ETPs, whereas Cxcr4 expression was lower (Fig. 7c). Flow cytometric analyses established surface expression of CCR6, CCR7 and CCR9 in E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs (Supplementary Fig. 6a). Furthermore, single cell analysis showed a high degree of co-expression of multiple Ccr genes in E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs (Supplementary Fig. 6b). Of further note, we found high expression of multiple paired immunoglobulin-like receptors (PIRs) in E11.5 T-IPs, which co-expressed lymphoid and GM genes but not MkE lineage-affiliated genes (Supplementary Fig. 6c,d). Contrary to what has previously been suggested42, Pir genes were almost undetectable in neonatal and adult ETPs, but sustained in E12.5 ETPs. While E11.5 HSPCs did not express any Pir genes, they were clearly upregulated in the E11.5 FL (but not adult) LMPPs, although at lower levels than in T-IPs (Supplementary Fig. 6c). Interestingly, FACS analysis of PIRA/B protein expression in E11.5 LMPPs and T-IPs corroborated the gene expression results and furthermore demonstrated that PIRA/B expression was increased in E11.5 Lin−c-Kit+CD25−Flt3+ _Rag1_-GFP+ circulating progenitors, compared to FL LMPPs, and further increased in E11.5 T-IP and E12.5 ETPs which shared a Lin−c-Kit+CD25−Flt3+ _Rag1_-GFP+ cell surface phenotype (Supplementary Fig. 4g and 6e). The combined T and myeloid lineage potential was comparable in PIR− and PIR+ E11.5 CD45+Lin−c-Kit+CD25−Flt3+ circulating progenitors (Supplementary Fig. 6f). Thus, multiple Pir genes are co-expressed in E11.5 lympho-myeloid T-IPs.
Figure 7. E11.5 thymopoiesis-initiating progenitors express a distinct repertoire of chemokine receptors.
(a) PCA of normalized global gene-expression of E11.5 Lin−CD45loVE-Cad+c-Kit+ AGM HSPCs (_n_=3), E11.5 Lin−c-Kit+Flt3+IL-7Rα+ FL LMPPs (_n_=3), E11.5 CD45+Lin−c-Kit+CD25−Flt3+ T-IPs (_n_=6), E12.5 CD45+Lin−c-Kit+CD25−Flt3+ ETPs (_n_=3) , E13.5 CD45+Lin−c-Kit+CD25+Flt3− DN2s (_n_=3), and neonatal (NN; 1 week, _n_=3) and adult (8 weeks, _n_=3) Lin−CD4−CD8α−c-Kit+CD25−Flt3+ ETPs. Each biological replicate (100 cells) was from a different pool of mice.
(b) Heatmap and Venn diagram based on RNA sequencing data, representing genes up-regulated (>4-fold) in CD45+Lin−c-Kit+CD25−Flt3+ T-IPs and ETPs at indicated developmental stage, relative to HSPCs/HSCs at equivalent stages.
(c) Expression of chemokine receptor genes, shown as mean (s.d.) RKPM, in HSPCs/HSCs (_n_=3), LMPPs (_n_=3), and T-IPs/ETPs (_n_=3-6) at different developmental stages (LMPPs and HSPC/HSC were not investigated at E12.5). Significant differences between developmental stages within each population; *p<0.05, **p<0.01, ***p<0.001; 0, below detection level.
Canonical Notch signaling is not required for T-IP colonization of the E11.5 thymus rudiment or pre-thymic T lineage restriction
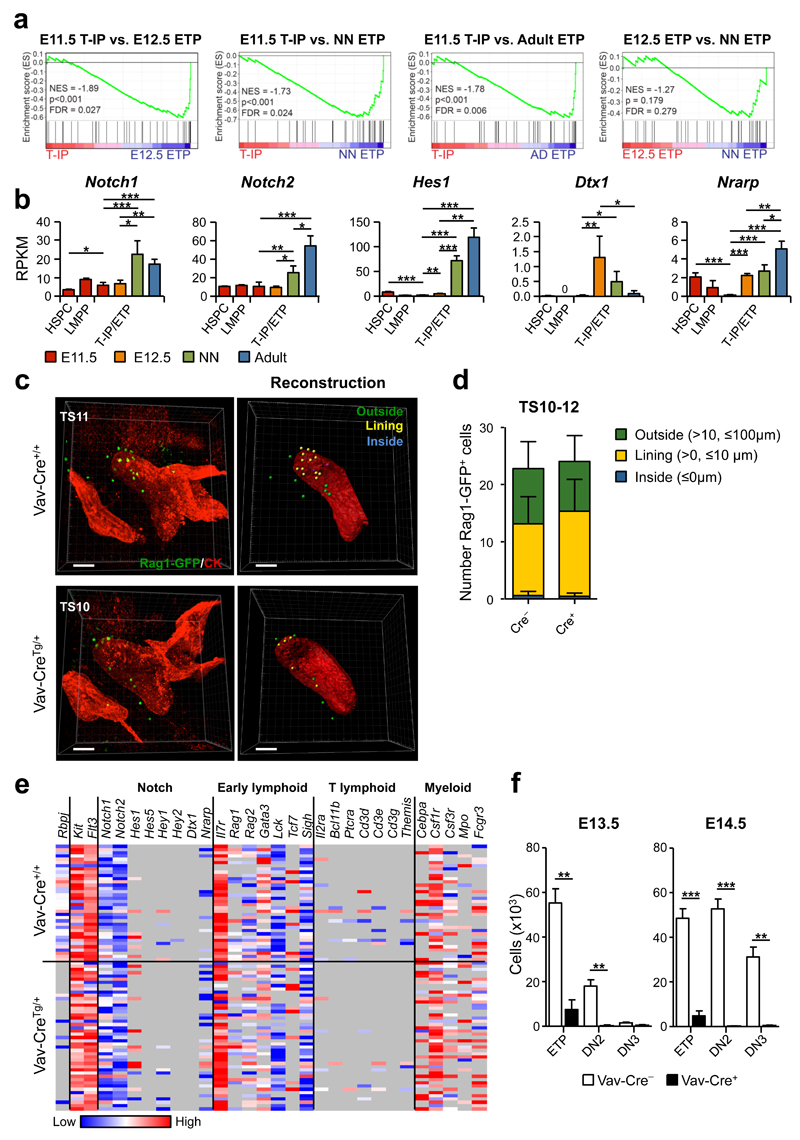
Previous studies suggested that embryonic T-IPs are activated by Notch-signaling already in the FL, resulting in pre-thymic T lineage restriction16, and other studies have implicated Notch signalling in the migration of BM TSPs to the thymus15. GSEA analysis of E11.5 T-IPs, E12.5, neonatal and adult ETPs, using a previously published gene set for Notch-signaling43 showed a marked enrichment of Notch-pathway related genes already in E12.5 ETPs compared to E11.5 T-IPs, and a less distinct upregulation from E12.5 to neonatal ETPs (Fig. 8a). A number of established Notch target genes, including Hes1, Deltex 1 (Dtx1) and Nrarp, were already clearly upregulated in E12.5 ETPs, but showed very low expression in E11.5 T-IPs, notably at similar or even lower levels than in the E11.5 FL LMPPs and HSPCs (Fig. 8b). Whole mount imaging analysis of TS10-12 embryos (Supplementary Fig. 7a-e) in which hematopoietic cells had been deleted for expression of Rbpj12, essential for canonical signaling through all Notch receptors13, demonstrated that the number of _Rag1_-GFP+ progenitors outside, lining and inside the thymus rudiment were unaffected in _Rbpj_-deficient TS10-12 embryos (Fig. 8c,d and Supplementary Fig. 7f,g), establishing that T-IP migration and initial seeding of the embryonic thymus at E11.5 occurs independently of canonical Notch-signaling.
Figure 8. Initial colonization of the E11.5 thymus rudiment takes place independently of Notch signaling.
(a) GSEA for Notch pathway genes (see Online Methods). NES, normalized enrichment score; FDR, false discovery rate (_n_=3 biological replicates per population).
(b) Notch receptor and target gene expression, shown as mean (s.d.) RPKM (_n_=3 per population). Significant differences between T-IPs/ETPs at different developmental stages; *p<0.05, **p<0.01, ***p<0.001; 0, below detection level. HSPCs and LMPPs were only analyzed at E11.5.
(c,d) Whole mount imaging of E11.5 (TS10-12) thymus colonization by _Rag1_-GFP+ (green) T-IPs in _Rbpj_Fl/Fl_Vav_-CreTg/+_Rag1_-GFPTg/+ (_Vav_-CreTg/+) and _Rbpj_Fl/Fl_Vav_-Cre+/+_Rag1_-GFPTg/+ (_Vav_-Cre+/+) littermate controls. (c) Three-dimensional image (left) and corresponding reconstruction (right) of the thymus rudiment. _Rag1_-GFP+ cells (right) are color coded according to their relative distance to the rudiment surface: Inside (blue) 0 µm; Lining (yellow), >0 and ≤10 µm; Outside (green), >10 and ≤100 µm. (d) Total number of cells inside, lining and outside the thymus rudiment, shown as mean (s.d.) representing 6 and 8 thymic lobes from _Rbpj_Fl/Fl_Vav-_CreTg/+_Rag1_-GFPTg/+ and _Rbpj_Fl/Fl_Vav-_Cre+/+_Rag1_-GFPTg/+, respectively. Thymus rudiment is marked by cytokeratin (CK, red). Scale bars represent 50 µm.
(e) Heatmap representation (ΔCt values, relative to Hprt) of Notch-related and lineage-affiliated genes in single E11.5 CD45+Lin−c-Kit+CD25−Flt3-T-IPs from _Rbpj_Fl/Fl_Vav_-CreTg/+_Rag1_-GFPTg/+ (_n_=46 cells from 6 embryos) and _Rbpj_Fl/Fl_Vav_-Cre+/+_Rag1_-GFPTg/+ (_n_=36 cells from 8 embryos) littermate controls from 2 litters. Only cells amplifying Flt3, c-Kit and Hprt (corresponding to 98% of total cells) were included. Grey indicates not detected.
(f) Impact of Rbpj-deficiency on thymocyte progenitor development. Mean (s.d.) absolute numbers of ETPs, DN2s and DN3s from E13.5 and E14.5 _Rbpj_Fl/Fl_Vav_-CreTg/+_Rag1_-GFPTg/+ (_Vav_-CreTg/+; _n_=3 and n=4, respectively) and _Rbpj_Fl/Fl_Vav_-Cre+/+_Rag1_-GFPTg/+ (_Vav_-Cre+/+; _n_=7 and n=11, respectively) littermate controls. **p<0.01, ***p<0.001.
Single cell gene expression analysis confirmed efficient _Rbpj_-deletion in T-IPs (Fig. 8e and Supplementary Fig. 8a). Since most Notch target genes can also be regulated through Notch-independent mechanisms, we hypothesized that the low expression of Notch target genes seen in E11.5 T-IPs might be independent of canonical Notch activation. In agreement, we found no difference in Notch target gene expression in single E11.5 T-IPs from _Rbpj_-deficient and WT littermate control embryos (Fig. 8e and Supplementary Fig. 8b). Moreover, the transcriptional lympho-myeloid lineage priming of T-IPs and expression of genes associated with T-cell lineage restriction was also unaffected by _Rbpj_-deficiency (Fig. 8e and Supplementary Fig. 8c,d), suggesting that canonical Notch signalling has no role in pre-thymic lineage restriction of T-IPs. Unlike the unperturbed T-IP colonization of the thymus rudiment, subsequent T-cell progenitor development was blocked at the DN1/ETP (CD4−CD8−c-Kit++CD25− or CD4−CD8−CD44+CD25-) stage already in E13.5 and E14.5 _Rbpj_-deficient embryos (Fig. 8f and Supplementary Fig. 8e,f), in agreement with the block reported later in embryonic development44 and in the adult thymus13. This strongly supports that the very earliest intrathymic T-cell progenitor progression and lineage restriction of embryonic T-IPs is critically dependent on canonical Notch-signaling, and in further agreement with this, E11.5 _Rbpj_Fl/Fl_Vav_-CreTg/+ thymic lobes with T-IPs were unable to progress beyond the ETP/DN1 (CD4−CD8−CD44+CD25−) stage on stromal cells expressing the Notch ligand delta-like 1 (DL1) (Supplementary Fig. 8g).
Collectively, these data demonstrate that unlike its critical role in intra-thymic T-lineage restriction canonical Notch signaling, is not required for the initial migration and seeding of the E11.25 embryonic thymus by _Rag1_-GFP+ T-IPs, nor in pre-thymic lineage-restriction of T-IPs.
Discussion
Herein we pursued studies to identify and molecularly and functionally characterize at the single cell level, the TSPs seeding the thymus rudiment for the first time during embryonic development (here named T-IPs), responsible for initiation of thymopoiesis and thereby contributing to the timely establishment of a competent immune system. The first embryonic wave of progenitors colonizing the thymus has for instance been shown to be responsible for the generation of a population of long-lived dendritic epithelial Vγ3+ T-cells, still present in the skin of adult mice17. In contrast to most previous studies, we purified and imaged T-IPs prior to the thymus being vascularized or colonized, providing a unique opportunity to visualize and characterize T-IPs/TSPs, rather than ETPs potentially already altered in their lineage-fate options following thymus-entry and Notch activation. This is critically important, as previous studies focused primarily on characterization of ETPs at E12.5 or later16, 17, 24, 45, but already at E12.5, Dll4 was highly expressed in a now ETP-occupied thymus, and Notch target genes upregulated in E12.5 ETPs, limiting studies to Notch-activated ETPs rather than pre-thymic TSPs.
Due to the narrow developmental timing in which the thymus anlage is seeded for the first time, we adapted a more accurate method for developmental staging through TS counting, establishing that colonization occurs at TS10-12. Subsequently, through imaging of _Vwf-_GFP embryos we established that fetal HSCs, which express _Vwf_-GFP at this stage31, do not appear to migrate to and seed the thymus at TS8-14. Moreover molecular analysis established that E11.5 HSPCs lack expression of chemokine receptors critical for migration to the embryonic thymus10, 41. This combined with absent megakaryocyte-erythroid and HSC potential in the E11.5 thymic rudiment strongly imply that AGM-derived HSCs are not responsible for the initial seeding of the E11.5 thymus rudiment. Rather, we found that the first TS10-12 T-IPs expressed _Rag1_-GFP and IL-7Ra, demonstrating that these (and other) lymphoid genes are uniformly turned on prior to T-IPs seeding the embryonic thymus rudiment. Importantly, this was established by imaging the initial _Rag1-_GFP+ T-IPs as they migrate through the surrounding mesenchyme to reach the thymus lining and ultimately the thymus rudiment. We also demonstrate that these T-IPs share the distinct Lin−c-Kit+CD25−Flt3+ _Rag1_-GFP+PIR+ phenotype with the first embryonic ETPs and can progress through the subsequent stages of thymic T-cell development and lineage restriction, generating ETPs, subsequent DN stages and CD4+CD8+ DP cells.
Although our findings show a close cell surface, molecular and functional relationship between E11.5 T-IPs and E11.5 FL PIRA/B+ LMPPs, we cannot rule out that T-IPs might also derive 32from the yolk sac, the hematopoietic site in which the first LMPPs appear to originate already at E9.532.
E11.5 T-IPs expressed distinct chemokine receptor genes. The increased expression of Ccr2, Ccr5, Ccr6 and Ccr7 in E11.5 T-IPs compared to neonatal and adult ETPs is in agreement with a previous study reporting expression of these genes in E12.5 CD45+ cells isolated from the perithymic mesenchyme46 and implies a different usage of chemokine receptors at this developmental stage. Together with the expression of known ligands for Ccr7, Ccr2 and Ccr5 in E14.5 thymic stromal cells47 this differential chemokine receptor expression pattern is likely to reflect the need of distinct migration properties for the colonization of the non-vascularized thymus rudiment. For instance, in agreement with a higher requirement for Ccr7 at pre-thymus rudiment vascularization stages, Ccr7 deficiency results in a stronger impairment of embryonic thymus colonization at early stages of embryonic development48, whereas Ccr2 and Ccr6 have yet to be functionally explored for a role in embryonic thymus colonization.
Whereas multiple previous studies suggested that embryonic TSPs represent largely T-cell-restricted progenitors lacking myeloid lineage potential16, 17, 24, 25, 35, our molecular and functional single cell studies establish that a large fraction of _Rag1-_GFP+IL-7Rα+ E11.5 T-IPs sustain combined myeloid and T-lineage programs and potential. Notably, progenitors with the same cell surface phenotype and combined T-myeloid lineage potential as T-IPs could be identified in the circulation of E11.5 embryos. While the finding of a fraction of T-IPs only reading out with T-cell potential in the OP9-DL1 assay is compatible with a fraction of T-IPs being T-cell-restricted prior to thymus entry, this might equally well reflect the inability of available assays to read out the short-lived myeloid lineage potential of T-IPs with 100% efficiency. Regardless, the high-resolution single cell molecular analysis demonstrating a lack of expression in T-IPs of genes uniformly expressed in the earliest T-cell-restricted progenitors in the embryonic thymus, including Ptcra, Cd3e and Bcl11b, failed to support previous claims that T-IPs contain T-cell-restricted progenitors. Also, expression of genes (Gata3 and Tcf7, also known as Tcf1) previously used to support that FL and blood might contain T-cell restricted progenitors35, are not only expressed in T-cell-restricted progenitors. Therefore, while it cannot be ruled out that T-IPs might include T-cell-restricted progenitors, evidence for this will require their prospective purification and characterization. While expression of PIRs has been suggested to mark a subset of T-cell-restricted FL progenitors42, E11.5 circulating PIR+ Lin−c-Kit+CD25−Flt3+_Rag1_-GFP+ progenitors possessed comparable frequencies of combined T-GM potential as Lin−c-Kit+CD25−Flt3+_Rag1_-GFP+ T-IPs, and single Pir+ T-IPs typically expressed combined lymphoid and myeloid programs.
The broad expression of PIR genes and protein might explain why most E11.5 T-IPs, like the PIR+ fraction of E11.5 FL LMPPs32 lack B-lineage potential, when compared to postnatal ETPs3, which we here show lack Pir expression. Nevertheless, consistent B-lymphoid lineage potential was detected in the dissected TS10-12 thymic rudiment at the same time-point as the T-cell potential. Since there are no B cell-restricted progenitors at this stage of development36, as confirmed here by fate mapping and molecular analysis, this strongly supports that T-IPs seeding the TS10-12 embryonic thymus include progenitors with combined T, B and myeloid, but immune-restricted, lineage potential.
The relevance of the myeloid lineage potential of adult ETPs remains unclear21. Given the rarity of T-IPs and what appears to be a rather restricted myeloid potential, this is unlikely to represent a significant pathway for extra-thymic myelopoiesis. However, since the _Rag1-_Cre fate mapping suggests that a large fraction of monocytes/macrophages in the embryonic thymus derive from TSPs, similar to granulocytes in the adult thymus20, TSPs might contribute locally towards generation of intra-thymic monocytes/macrophages suggested to play important roles in thymus development and homeostasis18, 19. It is also possible that the sustained myeloid program in TSPs primarily highlights that the final T-lineage-restriction step takes place in the thymus through loss of myeloid lineage potential.
Previous studies based on restricted lineage potentials and expression of canonical Notch target genes in candidate TSPs in FL and blood16, 17, proposed that pre-thymic Notch-activation might restrict the lineage potentials of TSPs prior to thymus colonization. However, our molecular and imaging analysis failed to provide any support for a role of canonical Notch-signaling in pre-thymic T-lineage restriction of T-IPs nor in their migration to the embryonic thymus. The lympho-myeloid lineage programming of T-IPs and the expression of genes associated with T-cell lineage restriction, were unperturbed in _Rbpj-_deficient T-IPs, even though deletion had already been induced at E9.5 in a pan-hematopoietic manner49. In agreement with these findings, we also found no evidence for canonical Notch activation when comparing Notch target gene expression in _Rbpj_-deficient and WT control E11.5 T-IPs. In contrast, subsequent intrathymic embryonic thymocyte progenitor progression and T-lineage restriction from T-IPs were strictly dependent on canonical Notch signalling.
In conclusion, through careful staging, imaging and single cell molecular analysis of the first embryonic progenitors migrating to and seeding the thymic rudiment, we demonstrate that this initial seeding occurs through lympho-myeloid-restricted T-IPs rather than T-cell restricted progenitors or HSCs as previously suggested. Moreover, while Notch-signaling has been proposed to play a role in pre-thymic T-IP lineage-restriction and migration, we provide evidence for a canonical Notch-independent embryonic pre-thymic processes of T-lineage development.
Online Methods
Mice
All mice were bred and maintained in accordance with UK Home Office regulations. Embryos were obtained from timed pregnancies, timed by the day of the vaginal plug, which was considered as E0.5. E11-11.75 embryos were additionally staged by tail somite counting (Supplementary Fig. 1). _Rag1_-GFP mice have been previously described3, 26 and were kindly provided by Dr N. Sakaguchi. To exclude maternal/placental cell contamination the _Rag1_-GFP transgene was always inherited from the male parent in plugging experiments. Cd79atm1(cre)Reth (_Mb1-_Cre) mice were kindly provided by Dr M. Reth37, _Rag1_-Cre mice by Dr. T. Rabbitts33 and vavP-Mcl1–trangenic mice were provided by S. Cory3. Both _Mb1-_Cre and _Rag1-_Cre strains were crossed with _R26_-stop-eYFP34 mice kindly provided by Dr S. Srinivas. To exclude maternal/placental cell contamination, the Cre or the _R26_-stop-eYFP alleles were never co-expressed in the same parent. _Vwf_-eGFP BAC transgenic mice31, _Rbpj_Fl/Fl mice12 _Vav_-Cre mice49 and W41 mice50 have been previously described. All mouse lines were backcrossed for at least 6 generations onto a CD45.2 C57BL/6 genetic background.
Dissection of thymic lobes
Embryos between E11 and E11.75 were placed in petri dishes containing PBS-Ca2+Mg2+ and individually staged according to the criteria shown in Supplementary Figure 1, prior to bilateral dissection of the thymic lobes. E11.5 specifies tail somite stages between 10-15, unless specified otherwise. The thymic lobes are located in the third branchial arches and are lined by the aortic arches (Supplementary Fig. 1c-e). With the embryo lying on its side, the tip of a watchmaker forceps (BioTek Microsurgery, P-110-AUF) was inserted into the aortic arch lateral to the thymic lobe and lifted up. This step was repeated for the two aortic arches above and below the lobe, resulting in its separation from the surrounding tissue. Moreover, this opening of the blood vessels results in a decreased contamination by blood cells. With the embryo lying on its back, the third branchial arch was further lifted up and the forceps inserted below the thymic lobe which was then lifted up to sever its connection with the pharynx. The contra-lateral lobe was similarly removed. Contaminating tissues (mainly vascular cells) were carefully removed from the lobes (Supplementary Fig. 1e and 2a). Single cell suspensions were prepared by mechanical disruption of the lobes through a syringe coupled with a 27G needle.
Calculation of thymus equivalents for immunohistochemistry analyses
To estimate the total number of equivalent thymuses (2 thymic lobes) analyzed, we performed serial sections of whole embryos and calculated the number of sections that would encompass the entire two thymic lobes. Based on sagittal serial sectioning and analysis of whole embryos at TS9, between 48-72 sections (6-8 µm each) encompassing the whole thymic lobes were obtained. Thus, 72 sections were set as an embryonic thymus equivalent and used to calculate the number of equivalent thymuses analyzed as specified in Figures 2b and 3b.
Fetal blood isolation
For fetal blood isolation embryos with the placenta were harvested, individualized and washed in clean PBS without disturbing the yolk sac. Clamps were then placed between the placenta and the yolk sac in order to block circulation between the embryo and the placenta, and the placenta was removed. Embryos without the placenta were washed again to avoid maternal blood contamination, transferred to a new dish with clean PBS and the clamps were removed to collect blood from umbilical cord and vitelin arteries. After 10 min the PBS (with blood cells) was collected, filtered and centrifuged for further analysis.
Immunofluorescence
For paraffin sections, embryos were fixed in Bouin's solution (Sigma) overnight at 4 °C, dehydrated in gradient ethanol and Histoclear (National Diagnostics) and embedded in paraffin (Sigma). Sagittal sections (6-8 µm) of whole embryos were prepared. Sections from paraffin embedded embryos were incubated with target antigen retrieval buffer (Dako) and proteinase K (Dako) treated. All sections were blocked with 5% fetal bovine, donkey or/and goat serum and incubated overnight at 4 °C with primary antibodies (including anti-GFP antibodies) followed by detection with conjugated secondary antibodies (Supplementary Table 7). Non-specific binding of antibodies was assessed by staining with either isotype controls followed by secondary antibodies, or secondary antibodies alone, as well as using WT controls whenever GFP transgenic embryos were used. Sections were mounted with coverslips using ProLong Gold Antifade Reagent (Invitrogen). Images were acquired using a BioRad Radiance 2000 laser scanning confocal microscope. Whole mount imaging was performed as described51. Briefly, trimmed embryos were fixed in 4% paraformaldehyde (PFA) and dehydrated in methanol series. Rehydrated embryos were then blocked in 2% goat serum, 2% fetal bovine serum (FBS) and 0.2% bovine serum albumin (BSA) and stained overnight with primary antibodies at 4°C, followed by overnight secondary antibody staining at 4°C. Embryos were then dehydrated in methanol, rendered transparent with benzyl alcohol/benzyl benzoate (BABB) and imaged in a Zeiss LSM-780 confocal microscope. Image analysis was performed with Imaris 7.5.0 software (Bitplane).
For single cell immunofluorescence analysis, cells were FACS-sorted directly into 50ul medium with 10% FBS on cover slips pre-coated with Poly-L-Lysine (Sigma-Aldrich). Cells were fixed for 10 min at room temperature with 2% PFA. Cover slips were then mounted in Vectashield with DAPI (Vector Laboratories) and imaged with a Zeiss LSM-780 confocal microscope. A single Z-stack was acquired for each cell.
Flow cytometry
All cell sorting experiments were performed on a BD FACSAriaIIu cell sorter (BD Biosciences) as previously described3, with a mean cell sorting purity of 99±0.5 % (mean ± s.e.m.). Single cells were seeded by an automated cell deposition unit (ACDU) providing single cells in >99% of the wells, and no wells with more than 1 cell. Single cell deposition efficiency was confirmed with fluorescent beads in all experiments. FACS analyses were performed on a BD LSRII (BD Biosciences) as previously described3. Fluorescence-minus-one controls as well as negative populations were used as gate-setting controls. See Supplementary Table 8 for detailed antibody information. Subsequent data analyses were performed with the FlowJo analysis software (Flowjo, LLC).
In vivo reconstitution assays
Fetal thymocytes (CD45.2) were transplanted together with 300,000 unfractionated support BM cells (CD45.1) into lethally irradiated (2x450cGy) C57BL6 (CD45.1) recipient mice (>8 weeks), either intravenously or intrafemorally. In transplantations using homozygous W41 recipients (CD45.1), mice were irradiated with a single dose at 600 cGy. Peripheral blood analyses were conducted between 3 and 16-17 weeks post-transplantation as previously described3, 32. The minimum detection values were determined based on the number of total events acquired by FACS for total donor cells and for each lineage separately. Mice with total donor chimerism above threshold values (>0.003%) and donor-derived lineage chimerism to T cells (>0.02%), B cells (>0.02%), myeloid cells (>0.25)% and platelets (>0.02%) were considered reconstituted.
BM chimera experiments were performed by transplantation of a mixture of _Rag1_-Cretg/+ and _R26_-stop-eYFPFL/FL (1:1) CD45.2 BM cells into lethally irradiated CD45.1 recipient mice (2x450cGy). Thymocyte and monocyte/macrophage reconstitution and eYFP expression in thymic macrophages was assessed 3 weeks after transplantation.
Evaluation of in vitro lineage potentials
GM and Mk lineage potentials were analyzed as previously described3 (see Supplementary Table 9 for details on cell culture conditions). Unfractionated fetal thymocytes and control FL cells were plated in Megacult collagen-based media (StemCell Technologies) supplemented with appropriate cytokines (Supplementary Table 9). Mk colonies were evaluated after 7 days following Acetylthiocholiniodide (Sigma) staining according to manufacturer’s instructions. For evaluation of erythroid potential, unfractionated fetal thymocytes and control FL cells were seeded in complete methylcellulose (GF M3434; StemCell Technologies). Cultures were evaluated after 8 days using 2,7-diaminofluorene staining (DAF; Sigma) as previously described3. To evaluate B and T lymphoid as well as myeloid lineage potential unfractionated fetal thymocytes or sorted single cells were plated onto monolayers of OP9 or OP9-DL1 stromal cells (Supplementary Table 9). Combined T lymphoid and myeloid potential from sorted single cells was evaluated on OP9-DL1 stromal cells in the presence of cytokines (Supplementary Table 9). Myeloid cells in OP9 or OP9-DL1 stroma co-cultures were evaluated after 6-7 days by morphological analysis of May-Grünwald (Sigma) and Giemsa (Fluka) stained cytospins. B and T lymphoid potential was evaluated after 2 weeks by FACS.
RNA sequencing analysis
Samples for RNA sequencing (80-100 cells) were prepared and sequenced as previously described32. Reads were aligned using Bowtie against the murine transcriptome (mouse NCBI build37 Refseq transcripts). Non-uniquely mapped reads were discarded. Count number of reads for each transcript and reads per kilobase of a transcript per million mapped reads (RPKMs) were calculated and assigned to each transcript. Overlapping Refseq transcripts were collapsed giving the highest one expression value per gene loci. The RPKM values were used when investigating the expression of specific genes. The non-adjusted read counts for each gene was used for statistical calculation of global differential expression between the specified populations using the DESeq package. Genes, which were significant at an adjusted p-value equal to or less than 0.05 (calculated using a Benjamini-Hochberg adjusted p-value) were considered differentially expressed between populations. The method has been validated for sensitivity, specificity and transcript coverage down to the single cell level32.
Hierarchical clustering and PCA were performed in R statistical programming environment (www.r-project.org). The hclust function (with Euclidean distance and the complete method) was done on log-transformed RPKM values for the clustering analysis of 1,029 genes. The pvclust function was used for the bootstrap resampling hierarchical clustering by samples. The same set of genes was subjected to the prcomp function for the PCA analysis. This set of genes was selected from genes that show high variation of expression levels among populations (adjusted p-values from rowFtests function equal to or less than 0.01 and coefficient of variation (CV) equal to or larger than 0.5) combining with the published gene sets for CLPs, Pre-MegEs3 and HSCs38. GSEA was performed using GSEA software (http://www.broadinstitute.org/gsea) with permutation on the gene sets, 1000 permutations, and log2 ratio of classes as metric for ranking genes. Gene sets used in this study have been previously published3, 38, 43. The number of genes up-regulated in T-IPs and ETPs was selected based on mean expression values equal to or larger than 4-fold change relative to all HSPCs/HSCs. These filtered genes were classified into categories (E11.5 T-IPs, NN ETP and adult ETPs) using the combination of present or absent logic operation according to the comparison between T-IPs/ETPs and HSPCs/HSCs. Pathways and Gene ontology analysis was done using the Pathway and GeneGO tool of MetaCore from Thomson Reuters (Version 6.15). RNA sequencing data sets for E11.5 LMPPs have previously been published32
Quantitative single cell PCR
Single cells were sorted directly into RT/pre-amplification mix using an automated cell deposition unit (ACDU). Multiplex quantitative PCR analysis of single E11.5 ETPs (BioMark 48.48 Dynamic Array platform; Fluidigm) using TaqMan Gene Expression Assays (Applied Biosystems) were performed as previously described3, 32. TaqMan assays used for the analysis are shown in Supplementary Table 10. Data were analyzed using the ΔCt method, using the house-keeping gene Hprt for normalization. Heatmap was generated using GENE-E software (http://www.broadinstitute.org/cancer/software/GENE-E/). For Dll4 expression analysis, 50-200 cells were sorted directly into Cells Direct 2X reaction mix and analyzed as previously described3. A no RT control was included and samples were run in triplicates using BioMark 48.48 Dynamic Array platform (Fluidigm) and mouse Universal Probe Library assays (Roche). Primers and probes used are described in Supplementary Table 11. A geometric mean of average Ct values for three housekeeping genes (Actb, Hmbs and Tbp) was used as reference Ct value. Data are shown as 2-ΔCt.
Statistical analysis
Statistical significance was determined by two-sided t-Test or two-sided Fisher’s exact test as indicated.
Supplementary Material
Supplementary Tables 1-6
Supplementary Information
Aknowledgments
We thank N. Sakaguchi (Kumamoto University) for providing the _Rag1_-GFP mouse line; M. Reth (Max Planck Institute of Immunobiology) for Cd79atm1(cre)Reth mice; S. Srinivas (University of Oxford) for _R26_-stop-eYFP mice; T. Rabbitts (University of Oxford) for _Rag1_-Cre mice; N. Iscove (Ontario Cancer Institute, University Health Network) for W41 mice; A. Cumano (Institut Pasteur) for OP9 and OP9-DL1 stromal cells; Biomedical Services at Oxford University for animal support; B. Wu, for excellent technical assistance; P. Sopp, S-A. Clark from the WIMM FACS facility (supported by the MRC HIU; MRC MHU (MC_UU_12009); NIHR Oxford BRC and John Fell Fund (131/030 and 101/517), the EPA fund (CF182 and CF170) and by the WIMM Strategic Alliance awards G0902418 and MC_UU_12025. C. Lagerholm and D. Waithe from the Wolfson Imaging Centre Oxford (supported by MRC via the WIMM Strategic Alliance (G0902418), the MHU (MC_UU_12009), the HIU (MC_UU_12010), the Wolfson Foundation (Grant 18272), by an MRC/BBSRC/EPSRC grant (MR/K015777X/1) to MICA – Nanoscopy Oxford (NanO): Novel Super-resolution Imaging Applied to Biomedical Sciences, Micron (107457/Z/15Z) and the WIMM Strategic Alliance awards G0902418 and MC_UU_12025. This work was supported by a programme grant (G0801073) and Unit Grant (MC_UU_12009/5) from the Medical Research Council UK (S.E.W.J.), an international recruitment grant from the Swedish Research Council (S.E.W.J.), Knut och Alice Wallenberg Foundation (WIRM; S.E.W.J.), StratRegen, KI (S.E.W.J.), EU-FP7 EuroSyStem Integrated projects (S.E.W.J., C.C.B.), the Hemato-Linné grant (Swedish Research Council Linnaeus; S.E.W.J.), the MRC Molecular Haematology Unit core award (M.d.B), EMBO-LTF (T.C.L.), Kay Kendall Leukaemia Fund Junior research fellowship (T.C.L.) and the Leukaemia and Lymphoma Research (C.C.B and A.J.M.).
Footnotes
Author Contribution
S.E.W.J., S.L. and T.C.L. conceptualized and designed the overall research, analyzed the data and wrote the manuscript, which was subsequently reviewed and approved by all authors. S.L. and I.G. established and performed embryonic staging experiments; T.C.L., S.L., H.B., I.G. and M.d.B. performed dissections; T.C.L, T.M. H.B. and E.A. did imaging analyses; P.S.W. processed RNA samples for RNAseq; S.T., T.C.L., I.C.M. and C.B. analyzed RNA seq data; S.L., T.C.L. and P.S.W. performed in vivo transplantations; S.L., T.C.L., M.L. and T.B.J. carried out in vitro lineage potential analyses; T.C.L., H.V., A.J.M. and D.A. did single cell and quantitative PCR analyses; S.L., T.C.L. and J.C. carried out flow cytometry experiments; T.C.L. did fate mapping analyses; A.J.M., T.C.L., S.L. and H.B. performed morphology analyses; R.P.,F.G., C.N., R.S., M.d.B., C.C.B. and I.G. contributed with advice and input on experimental design and data analysis.
Competing Financial Interests
The authors declare no competing financial interests.
Accession codes.
GEO: RNA sequencing data, GEO76140
References
- 1.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 3.Luc S, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 5.Jenkinson EJ, Jenkinson WE, Rossi SW, Anderson G. The thymus and T-cell commitment: the right niche for Notch? Nat Rev Immunol. 2006;6:551–555. doi: 10.1038/nri1883. [DOI] [PubMed] [Google Scholar]
- 6.Mori S, Shortman K, Wu L. Characterization of thymus-seeding precursor cells from mouse bone marrow. Blood. 2001;98:696–704. doi: 10.1182/blood.v98.3.696. [DOI] [PubMed] [Google Scholar]
- 7.Ciofani M, Zuniga-Pflucker JC. The thymus as an inductive site for T lymphopoiesis. Annu Rev Cell Dev Biol. 2007;23:463–493. doi: 10.1146/annurev.cellbio.23.090506.123547. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn CC, Manley NR. Developing a new paradigm for thymus organogenesis. Nat Rev Immunol. 2004;4:278–289. doi: 10.1038/nri1331. [DOI] [PubMed] [Google Scholar]
- 9.Owen JJ, Ritter MA. Tissue interaction in the development of thymus lymphocytes. J Exp Med. 1969;129:431–442. doi: 10.1084/jem.129.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, et al. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 11.Ferrero I, et al. DL4-mediated Notch signaling is required for the development of fetal alphabeta and gammadelta T cells. Eur J Immunol. 2013;43:2845–2853. doi: 10.1002/eji.201343527. [DOI] [PubMed] [Google Scholar]
- 12.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 13.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 14.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 15.Yu VW, et al. Specific bone cells produce DLL4 to generate thymus-seeding progenitors from bone marrow. J Exp Med. 2015;212:759–774. doi: 10.1084/jem.20141843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harman BC, et al. T/B lineage choice occurs prior to intrathymic Notch signaling. Blood. 2005;106:886–892. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- 17.Ramond C, et al. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol. 2014;15:27–35. doi: 10.1038/ni.2782. [DOI] [PubMed] [Google Scholar]
- 18.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tacke R, et al. The transcription factor NR4A1 is essential for the development of a novel macrophage subset in the thymus. Sci Rep. 2015;5:10055. doi: 10.1038/srep10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Obaldia ME, Bell JJ, Bhandoola A. Early T-cell progenitors are the major granulocyte precursors in the adult mouse thymus. Blood. 2013;121:64–71. doi: 10.1182/blood-2012-08-451773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlenner SM, Rodewald HR. Early T cell development and the pitfalls of potential. Trends Immunol. 2010;31:303–310. doi: 10.1016/j.it.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Rae F, et al. Characterisation and trophic functions of murine embryonic macrophages based upon the use of a Csf1r-EGFP transgene reporter. Dev Biol. 2007;308:232–246. doi: 10.1016/j.ydbio.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 23.Peault B, Khazaal I, Weissman IL. In vitro development of B cells and macrophages from early mouse fetal thymocytes. Eur J Immunol. 1994;24:781–784. doi: 10.1002/eji.1830240345. [DOI] [PubMed] [Google Scholar]
- 24.Kawamoto H, Ohmura K, Katsura Y. Presence of progenitors restricted to T, B, or myeloid lineage, but absence of multipotent stem cells, in the murine fetal thymus. J Immunol. 1998;161:3799–3802. [PubMed] [Google Scholar]
- 25.Masuda K, et al. Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J Immunol. 2005;174:2525–2532. doi: 10.4049/jimmunol.174.5.2525. [DOI] [PubMed] [Google Scholar]
- 26.Yokota T, et al. Tracing the first waves of lymphopoiesis in mice. Development. 2006;133:2041–2051. doi: 10.1242/dev.02349. [DOI] [PubMed] [Google Scholar]
- 27.Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 28.Gordon J, Manley NR. Mechanisms of thymus organogenesis and morphogenesis. Development. 2011;138:3865–3878. doi: 10.1242/dev.059998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch U, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calderon L, Boehm T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell. 2012;149:159–172. doi: 10.1016/j.cell.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 31.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 32.Boiers C, et al. Lymphomyeloid Contribution of an Immune-Restricted Progenitor Emerging Prior to Definitive Hematopoietic Stem Cells. Cell Stem Cell. 2013;13:535–548. doi: 10.1016/j.stem.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 33.McCormack MP, Forster A, Drynan L, Pannell R, Rabbitts TH. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol Cell Biol. 2003;23:9003–9013. doi: 10.1128/MCB.23.24.9003-9013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikawa T, et al. Identification of the earliest prethymic T-cell progenitors in murine fetal blood. Blood. 2004;103:530–537. doi: 10.1182/blood-2003-06-1797. [DOI] [PubMed] [Google Scholar]
- 36.Delassus S, Cumano A. Circulation of hematopoietic progenitors in the mouse embryo. Immunity. 1996;4:97–106. doi: 10.1016/s1074-7613(00)80302-7. [DOI] [PubMed] [Google Scholar]
- 37.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers SM, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rybtsov S, et al. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J Exp Med. 2011;208:1305–1315. doi: 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zlotoff DA, et al. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda K, et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercher T, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3:314–326. doi: 10.1016/j.stem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chea S, et al. Single-Cell Gene Expression Analyses Reveal Heterogeneous Responsiveness of Fetal Innate Lymphoid Progenitors to Notch Signaling. Cell Rep. 2016;14:1500–1516. doi: 10.1016/j.celrep.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Desanti GE, et al. Clonal analysis reveals uniformity in the molecular profile and lineage potential of CCR9(+) and CCR9(-) thymus-settling progenitors. J Immunol. 2011;186:5227–5235. doi: 10.4049/jimmunol.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkinson WE, et al. Chemokine receptor expression defines heterogeneity in the earliest thymic migrants. Eur J Immunol. 2007;37:2090–2096. doi: 10.1002/eji.200737212. [DOI] [PubMed] [Google Scholar]
- 47.Liu C, et al. The role of CCL21 in recruitment of T-precursor cells to fetal thymi. Blood. 2005;105:31–39. doi: 10.1182/blood-2004-04-1369. [DOI] [PubMed] [Google Scholar]
- 48.Lu M, et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRbeta chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 49.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 50.Geissler EN, McFarland EC, Russell ES. Analysis of pleiotropism at the dominant white-spotting (W) locus of the house mouse: a description of ten new W alleles. Genetics. 1981;97:337–361. doi: 10.1093/genetics/97.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokomizo T, et al. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat Protoc. 2012;7:421–431. doi: 10.1038/nprot.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1-6
Supplementary Information