Mutations in PIH1D3 Cause X-Linked Primary Ciliary Dyskinesia with Outer and Inner Dynein Arm Defects (original) (raw)
Abstract
Defects in motile cilia and sperm flagella cause primary ciliary dyskinesia (PCD), characterized by chronic airway disease, infertility, and left-right body axis disturbance. Here we report maternally inherited and de novo mutations in PIH1D3 in four men affected with PCD. PIH1D3 is located on the X chromosome and is involved in the preassembly of both outer (ODA) and inner (IDA) dynein arms of cilia and sperm flagella. Loss-of-function mutations in PIH1D3 lead to absent ODAs and reduced to absent IDAs, causing ciliary and flagellar immotility. Further, PIH1D3 interacts and co-precipitates with cytoplasmic ODA/IDA assembly factors DNAAF2 and DNAAF4. This result has clinical and genetic counseling implications for genetically unsolved male case subjects with a classic PCD phenotype that lack additional phenotypes such as intellectual disability or retinitis pigmentosa.
Main Text
Motile cilia and flagella are microtuble-based organelles that extend from the surface of many cell types in the human body. Function of both flagella and cilia requires well-coordinated ATP-dependent interactions between the peripheral microtubuli and the axoneme.1 The basic structure of motile cilia consists of a ring of nine peripheral microtubule doublets surrounding one central pair (9+2 structure). The peripheral ring is connected to the central pair (CP) through radial spokes (RSs) and the nexin-dynein regulatory complex (N-DRC) connects adjacent doublets. The CP, N-DRC, and inner dynein arms (IDAs) are responsible for modulation and regulation of the ciliary movement while the outer dynein arms (ODAs) are responsible for beat generation.2, 3, 4, 5 ODAs and IDAs are large multimeric protein complexes that are pre-assembled in the cytoplasm before being transported to the axonemes.6, 7 The identification of proteins responsible for correct assembly, DNAAFs, and the composition of these protein complexes are critical to understand the patho-mechanisms of motile cilia-related diseases such as primary ciliary dyskinesia (PCD).
Primary ciliary dyskinesia (MIM: 244400) affects an estimated 1:15,000 live births and is characterized by abnormal ciliary and flagellar movement, leading to numerous severe health issues.8 Ineffective mucociliary clearance causes mucus stasis in the entire respiratory tract, leading to recurrent infections and chronic inflammation. Dysfunctional cilia at the embryonic node give rise to laterality defects such as situs inversus of the internal organs in about half of the individuals.9 In addition, PCD-affected individuals often suffer from sub- or infertility caused by dysfunction of fallopian tube cilia and sperm flagella.
PCD is a genetically heterogeneous ciliopathy owing to the large number of proteins that are involved in ciliary motility. So far, autosomal-recessive mutations in 30 genes account for an estimated 70% of cases.10 Genetic analyses of PCD-affected individuals identified several autosomal-recessive mutations in genes encoding for axonemal subunits of the ODA and ODA-docking complexes (ODA-DCs).11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 In addition, recessive mutations in genes encoding other components of the ciliary motility apparatus as well as proteins required for motile ciliogenesis have been identified to be disease causing. An overview of these genes is given by Werner and colleagues.26 Among those are also genes encoding proteins involved in cytoplasmic pre-assembly of ODA and IDA that have emerged from mutation analyses of PCD-affected individuals: DNAAF1 (LRRC50 [MIM: 613190]),27, 28 DNAAF2 (KTU [MIM: 612517]),29 DNAAF3 (C19orf51 [MIM: 614566]),30 DNAAF4 (DYX1C1 [MIM: 608709]),31 DNAAF5 (HEATR2 [MIM: 614864]),32 LRRC6 (MIM: 614930),33 ZMYND10 (MIM: 607070),34, 35 SPAG1 (MIM: 603395),36 and C21ORF59 (MIM: 615494).37 Additionally, two X-linked PCD variants have been reported and are associated with syndromic cognitive dysfunction or retinal degeneration caused by mutations in OFD1 (MIM: 311200) and RPGR (MIM: 312610), respectively.38, 39 Here, we describe another example of X-linked non-syndromic PCD caused by mutations in PIH1D3.
We assembled a cohort of 75 PCD-affected individuals. Diagnosis of PCD was based on a combination of clinical symptoms and examination of ciliary motion by high-speed videomicroscopy (HSVM), ciliary ultratructure by transmission electron microscopy (TEM), and nasal nitric oxide production rate. We first excluded individuals from the Volendam population carrying a previously identified CCDC114 founder mutation.22
We screened these 75 PCD-affected individuals with a targeted-exome panel of 310 genes. This panel included 26 PCD-associated genes and a subset of candidate genes. These candidate genes were selected based on (1) at least 10-fold higher gene expression via RNA microarray (data not shown) during in vitro ciliogenesis of human airway epithelium cells, and (2) a previous association to ciliary or flagellar proteins or processes in published candidate gene lists and the Gene Network database. Signed and informed consent was obtained from all individuals fulfilling the diagnostic criteria of PCD and their family members, according to protocols approved by the Institutional Ethics Review board at the VU University Medical Center (Amsterdam, the Netherlands) and by the Institutional Ethics Review board at the University of Muenster (Muenster, Germany). Targeted-exome sequencing of genomic DNA was performed at the VU University Medical Center and at the Cologne Center for Genomics (CCG). For enrichment, the Truseq DNA LT Sample prep kit or the NimbleGen SeqCap EZ Human Exome Library v.2.0 was used. Enriched preparations were sequenced with the HiSeq2000 platform or the HiSeq2500 (Illumina) as paired end 2 × 100 base pairs reads. The 30× coverage reached 92.4%. Sequencing reads that passed quality filtering were mapped to the reference genome sequence (hg19). Variants that were present in dbSNP, the 1000 Genomes Project, and Exome Aggregation Consortium (ExAC) with a minor allele frequency > 0.01 were excluded. We focused on nonsynonymous mutations, splice-site substitutions, and indels following an autosomal-recessive and X-linked inheritance pattern.
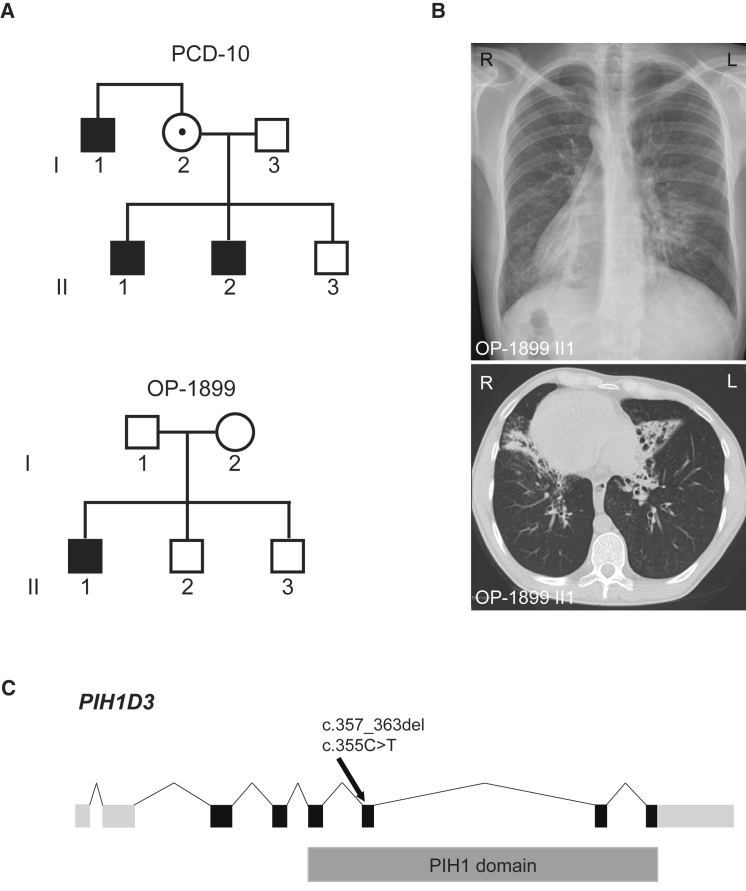
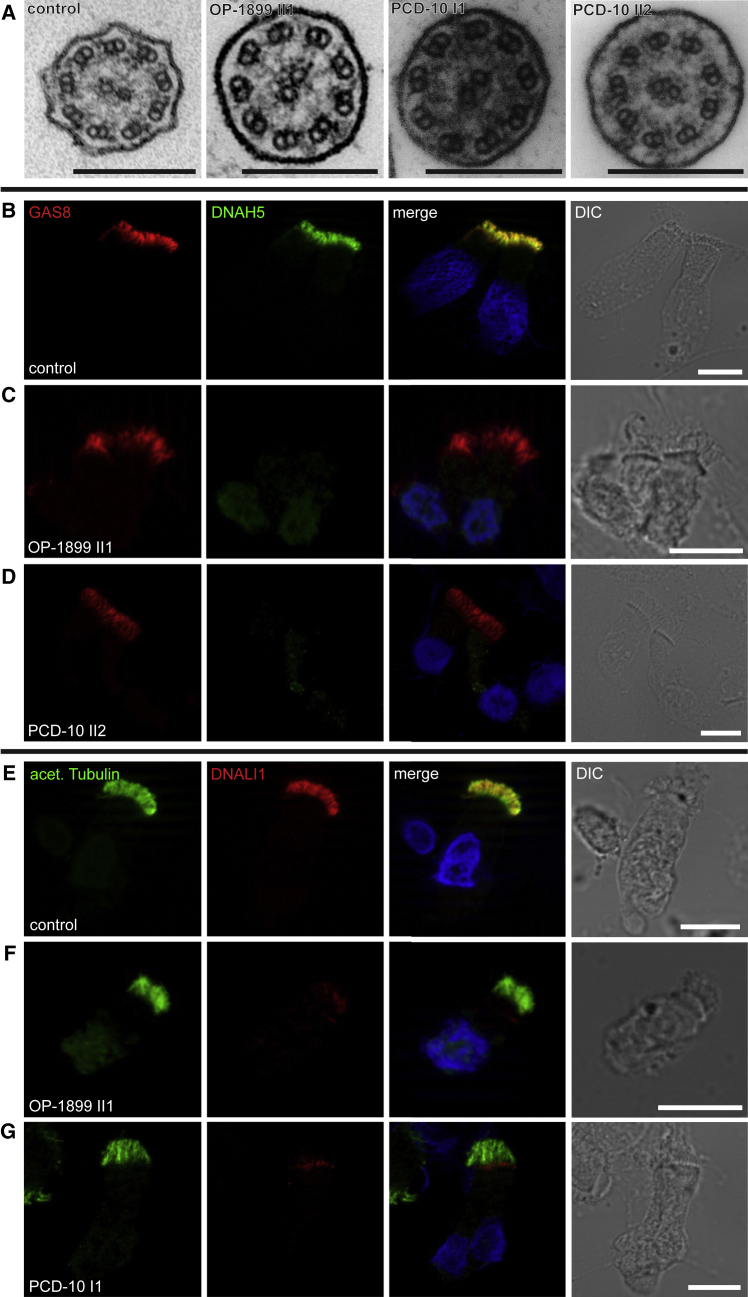
This approach revealed a hemizygous mutation in PIH1D3 (GenBank: NM_001169154.1, c.357_363del [p.Val120Leufs∗6]) in PCD-10 II-1 (Figure 1A) and a hemizygous nonsense mutation (c.355C>T [p.Gln119∗]) within exon 6 of PIH1D3 OP-1899 II-1 (Figure 1A). Neither mutation is reported in genomic variant databases such as dbSNP, 1000 Genomes, the Exome Variant Server (EVS), or the ExAC database. Both affected individuals show classical PCD symptoms such as chronic sinusitis, chronic otitis media, and chronic lower respiratory tract infections as well as bronchiectasis in the middle lobe and mucus plugging (shown for OP-1899 II-1 in Figure 1B). In addition, both have situs inversus totalis (shown for OP-1899 II-1 in Figure 1B) and neonatal respiratory distress syndrome, a typical finding often present in PCD. Nasal nitric oxide measurement, high-speed videomicroscopy in sequential monolayer-suspension cell culture, and transmission electron microscopy (TEM) was performed on all individuals of the PCD-10 family as previously described.22 Nasal NO production rate was measured either by using a hand-held Niox Mino (Aerocrine) or EcoMedics CLD88 (Duernten) and was low in all affected individuals (Table 1). Careful analysis of cilia from PCD-10 I-1, PCD-10 II-1, PCD-10 II-2, and OP-1899 II-1 showed complete immotilite respiratory cilia (Movies S1 and S2) when compared to control subjects (Movies S3 and S4). OP-1899 II-1 showed complete immotile sperm flagella (Movie S5) when compared to control subjects (Movie S6). The motility findings in respiratory cilia and sperm flagella are consistent with a loss of ODA. Additionally, PCD-10 II-1 and PCD-10 II-2 had undergone fertility testing in the past, which showed largely immotile sperm flagella. TEM analyses of respiratory cilia of all affected showed absence of ODAs and a possible reduction of IDAs (Figure 2A).
Figure 1.
Results of the Mutational Analyses in PIH1D3, Pedigrees of the PCD-Affected Families PCD-10 and OP-1899, and Clinical Features of OP-1899 II1
(A) Hemizygous loss-of-function mutations in PIH1D3 located on the X chromosome were identified in two families. Pedigree of PCD-affected families PCD-10 and OP-1899. PCD-affected siblings are shaded black and the unaffected sibling is shaded white.
(B) The chest X-ray radiograph as well as two computed tomography scans show situs inversus totalis, chronic airway disease with bronchiectasis in the middle lobe, and mucus plugging in OP-1899 II1.
(C) Exon-intron structure of PIH1D3. The exon-intron structure of PIH1D3 with untranslated (light gray) and translated (black) regions and the PIH1 domain (dark gray).
Table 1.
Diagnostic Characteristics
| Affected Individuals | TEM Defect | HSVM | nNO (nL/min) | Nasal Congestion | Neonatal Period | SI | Chronic Otitis Media | Recurrent Respiratory Infections | Chronic Wet Cough | Fertility Defect |
|---|---|---|---|---|---|---|---|---|---|---|
| PCD-10 I:1 | ODA | immotile | 2 | yes | unknown | no | yes | yes | yes | yes, 2 adopted children |
| PCD-10 II:1 | not enough cilia | immotile | 5 | yes | pneumonia | yes | yes | yes | yes | yes |
| PCD-10 II:2 | ODA | immotile | 4 | yes | pneumonia | no | yes | yes | yes | yes |
| OP-1899 II1 | ODA | immotile | 38 | yes | respiratory distress syndrome | yes | yes | yes | yes | yes |
Figure 2.
Loss-of-Function PIH1D3 Mutations Result in Absence of Outer and Inner Dynein Arms in Respiratory Epithelial Cells
(A) The analysis of transmission electron microscopy cross-sections from respiratory cilia from OP-1899 II1, PCD-10 I1, and PCD-10 II2 showed a normal 9+2 architecture with absence of outer dynein and inner dynein arms. Scale bars represent 200 nm.
(B–D) Respiratory epithelial cells from control (B) and PCD-affected individuals OP-1899 II1 (C) and PCD-10 II2 (D) were double-labeled with antibodies directed against outer dynein arm heavy chain DNAH5 (green) and the N-DRC component GAS8 (red). Both proteins colocalize along the cilia in cells from the unaffected controls (B, yellow). In contrast, in mutant cells, DNAH5 was absent from or severely reduced in ciliary axonemes (C and D). The green staining in the nuclei observed in (C) is most probably background.
(E–G) Cells were double-labeled with antibodies directed against acetylated tubulin (green) and the inner dynein arm light chain DNALI1 (red). In unaffected control cilia, both proteins colocalize along the ciliary axonemes (E, yellow). In contrast, in cells of individuals carrying PIH1D3 mutations, DNALI1 was absent from or severely reduced in the ciliary axonemes (F and G). The red staining at the ciliary base is an artifact caused by the polyclonal rabbit antibody.46 Nuclei were stained with Hoechst33342 (blue). Scale bars represent 10 μm.
To explore the possibility of an X-linked recessive inheritance mode in both families, we investigated the pedigrees (Figure 1A). Individual PCD-10 II-1 has a brother (PCD-10 II-2) and uncle (PCD-10 I-1) with similar respiratory symptoms and a history of sub- or infertility (Table 1). Sanger sequencing confirmed the mutation in PCD-10 I-1, PCD-10 II-1, and PCD-10 II-2 and in one of the alleles of the mother of individuals PCD-10 II-1 and PCD-10 II-2, confirming an X-linked recessive inheritance mode (Figure 1A). Pedigree analysis of family OP-1899 shows that OP-1899 II-1 has two unaffected brothers who did not carry the hemizygous mutation. Segregation analyses in family OP-1899 revealed that this is a de novo mutation. We sequenced PIH1D3 in an additional 40 male PCD-affected individuals with ODA or combined ODA/IDA defects for mutations in PIH1D3 but did not identify additional mutations.
PIH1D3 (also known as CXorf41) is located on chromosome Xq22.3, comprises 8 exons, and encompasses approximately 38 kb of genomic DNA. PIH1D3 is one of four members of a protein family interacting with Hsp90 (PIH1). Studies in Chlamydomonas suggest that PIH1 functions in the preassembly of axonemal dyneins but may also function in preribosomal RNA processing.40, 41 Mutations in DNAAF2/KTU, also containing a PIH1 domain, cause PCD by interferring with cytoplasmic preassembly of both ODAs and IDAs in respiratory cilia and sperm flagella.29 Further, PIH1D3 mouse and zebrafish orthologs have previously been linked to flagellar motility defects and polycystic kidney disease, respectively.41, 42 _Pih1d3_−/− mice almost completely lack sperm ODAs and show a reduced number of IDAs. The mutations we identified in both families lie in the crucial PIH1 domain (Figure 1C) and are expected to cause either a truncated protein or no protein at all as a consequence of nonsense-mediated mRNA decay (NMD).
To assess the functional impact of the c.357_363del mutation, we synthesized cDNA from isolated RNA of respiratory ciliated cells of PCD-10 I-1, PCD-10 II-1, and PCD-10 II-2 and a healthy control subject. RNA was isolated after sequential monolayer-suspension cell culture, as described previously.22 PCR amplification of cDNA was carried out with primers for exons 5 and 7 of PIH1D3. PCR amplified one transcript in wild-type cells and two transcripts in mutant cells (Figure S1A). Sequence analysis of mutant mRNA revealed a transcript that includes the c.357_363del in exon 6 and an alternative splice product that skips exon 6. Each of these transcripts alters the reading frame to introduce a premature termination in exon 6 and exon 8, respectively, resulting in truncated proteins of 125 amino acids (c.357_363del) and 181 amino acids (alternative splice product), respectively. This is in contrast to the 214 amino acid full-length protein.
To determine whether NMD degrades these transcripts, we incubated respiratory cells of individual PCD-10 I-1 with 0.25 mg/mL cycloheximide for 4.5 hr to stabilize the RNA by blocking protein translation.22 qPCR of cDNA from respiratory cells of PCD-10 I-1 with and without cycloheximide incubation showed a 76% reduced expression of PIH1D3, relative to housekeeping gene VCP, in cells without cycloheximide (Table S1). Sequence analysis (Figure S1A) showed that while the two aberrant transcripts are present in equal proportion in cells without cycloheximide incubation, the transcript containing the c.357_363del is produced predominantly in cells with cycloheximide incubation. The transcript that skips exon 6 is probably caused by loss of exonic splice enhance sites and produces p.Tyr112Leufs∗32. This transcript terminates in the last exon, presumably rendering the transcript insensitive to NMD. The transcript containing the c.357_363del results in p.Val120Leufs∗6 but is subject to NMD, as was shown in cDNA from cells cultured with or without cycloheximide (Figure S1A). Both of these transcripts, if translated, will lead to severely altered proteins that are functionally inactive. The c.355C>T mutation in individual OP-1899 II-1 introduces a stop codon just one triplet before the c.357_363del variant does and it is therefore expected to cause a similar effect.
Next we performed western blot analysis to investigate the effect of the c.357_363del mutation at the protein level. Nasal epithelial cells were lysed in NuPAGE LDS Sample Buffer with NuPAGE reducing agent to produce a whole cell lysate, which was subjected to gel electrophoresis in NuPAGE 4%–12% BT gels using the XCell SureLock electrophoresis system. Proteins were transferred to nitrocellulose membrane using the iBlot Dry Blotting system (Invitrogen) and blocked in Odyssey blocking buffer (Westburg). The membrane was incubated overnight in Odyssey blocking buffer with 0.1% Triton X-100 with primary antibodies against PIH1D3 and actin. Details of the antibodies used are shown in Table S2. Secondary antibody incubation was performed for 1 hr with the IRDye 800 CW goat anti-rabbit IgG and the IRDye 680 CW goat anti-mouse IgG antibodies. The Odyssey infrared imaging system equipped with the Odyssey v.4 software (LI-COR Biosciences) was used to visualize fluorescence. Examination of respiratory epithelial cell lysates from individuals PCD-10 I-1, PCD-10 II-1, and PCD-10 II-2 failed to detect PIH1D3 in contrast to the healthy control (Figure S1).
To further characterize the ultrastructural defect caused by loss of function of PIH1D3, we performed high-resolution immunofluorescence (IF) analyses of control and PIH1D3 mutant respiratory cilia and sperm flagella as previously reported.12, 22 High-resolution IF images were taken with a Zeiss Apotome Axiovert 200 or a Zeiss LSM880 and processed with AxioVision 4.8 or ZEN Black and Adobe CS4. Confirming the ODA defect previously observed by TEM, we found complete axonemal absence of the ODA heavy chains DNAH5 (Figure 2) and DNAH9 in mutant respiratory cilia (Figure S2) and complete loss of the ODA intermediate chains DNAI1 and DNAI2 in mutant respiratory cilia (Figure S3). Thus, PIH1D3 deficiency results in abnormal assembly of type 1 as well as type 2 ODA complexes.1, 43 Additionally, DNAI1 and DNAI2 were both absent from mutant sperm flagella (Figure 3), indicating that assembly of both ODA types is also disturbed in sperm cells, similar to findings in the _Pih1d3_−/− mice.17 Interestingly, the inner dynein arm light chain DNALI1 was greatly reduced or absent in the respiratory ciliary axonemes (Figures 2E–2G). These findings are consistent with a defective cytoplasmic preassembly of ODAs and IDAs. However, normal localization of GAS8 and CCDC114 along the mutant ciliary axonemes shows that the N-DRC and ODA-DCs are not affected by loss of function of PIH1D3 (Figures 2 and S4). In contrast to our finding, respiratory cells are not disrupted in _Pih1d3_−/− mice. This can probably be explained by a species difference. Mice have two copies of Pih1d3, on the X chromosome and on chromosome 1, whereas PIH1D3 in humans is located solely on the X chromosome.
Figure 3.
PIH1D3 Mutant Sperm Flagella Are Deficient for the Outer Dynein Arm Intermediate Chains DNAI1 and DNAI2
Sperms were double-labeled with antibodies directed against acetylated tubulin (green) and DNAI1 or DNAI2 (both in red). DNAI1 and DNAI2 colocalize with acetylated tubulin along the flagellum in sperms from unaffected controls (yellow) (A and C). In contrast, in PIH1D3 mutant sperm tails, DNAI1 (B) and DNAI2 (D) were absent from flagellar axonemes. Nuclei were stained with Hoechst33342 (blue). Scale bars represent 10 μm.
The function of human PIH1D3 has not been previously investigated and the precise function of mouse Pih1d3 has likewise remained largely elusive. Dong et al. hypothesized that PIH1D3 may play a role in dynein arm stabilization as they detected endogenous protein-protein interaction between PIH1D3 and the chaperones HSP70 and HSP90 as well as DNAIC2 in mouse sperm. However, interaction with DNAAF 1, 2, and 3 was not described.41
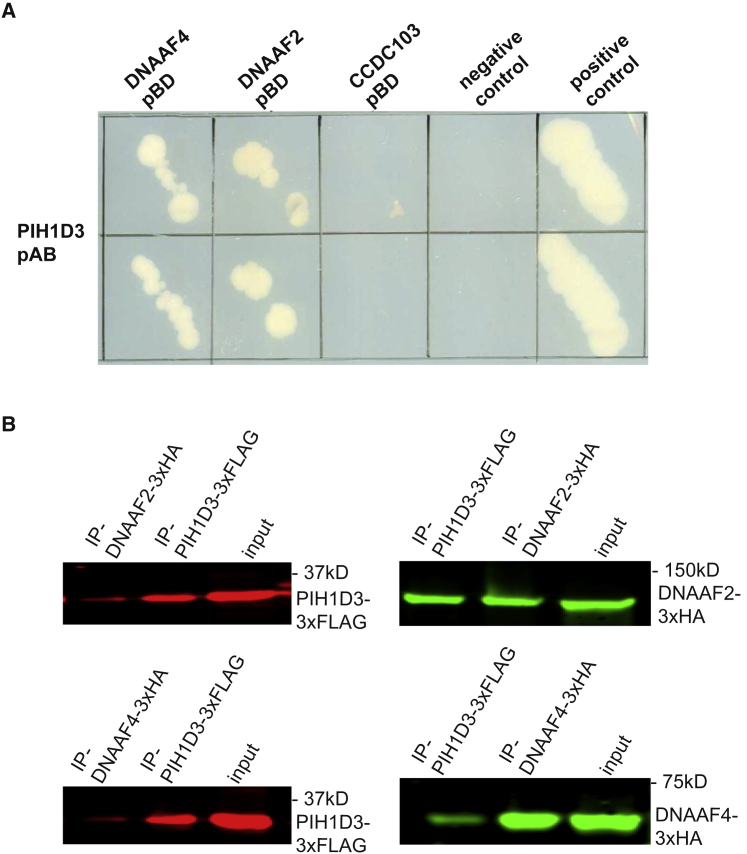
In order to better understand PIH1D3 function within this machinery, we performed a yeast two-hybrid (Y2H) screen using PIH1D3 as a bait to test interaction with several genes previously associated with PCD, including dynein arm assembly factors (DNAAFs), genes possibly involved in dynein arm transport (WDR69, IFT46), and others. Direct interaction between PIH1D3 and possible interactors was tested as previously described.31 Human PIH1D3 and HSP90 clones were purchased from Origene. All other cDNAs have been previously cloned by nested PCR from human bronchial epithelial cell cDNA (ScienCell) using KOD polymerase according to manufacturer’s directions as previously described.24 Due to its large size (making it unsuitable for Y2H screening), HSP90 cDNA was subcloned in fractions and similarly to all other cDNAs, recombined with pDONR201 Gateway vector via BP Clonase II reaction. Subsequently, cDNAs were subcloned into 3xHA and 3xFLAG epitope-tagged Gateway destination vectors via LR Clonase reaction. All cDNA clones were confirmed by sequence analysis and matched RefSeq gene accession numbers. HSP90 was used as a positive control and all tested genes are shown in Table S3. This led to the identification of two possible PIH1D3 interactors (Figure 4A): DNAAF2/KTU and DNAAF4/DYX1C1. To confirm these interactions on the protein level, we performed co-immunoprecipitations using overexpression of tagged versions of the proteins in HEK293T cells. Whole-cell extracts and western blots were conducted as previously described.44 Plasmids expressing C-terminal FLAG-tagged PIH1D3 were co-transfected with plasmids expressing C-terminal HA-tagged DNAAF2, DNAAF4, CCDC103, or p63 in HEK293T cells. At 24 hr after transfection, cells were lysed on ice in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40) supplemented with complete protease inhibitor cocktail (Roche Diagnostics). Lysates were incubated with either anti-FLAG M2-agarose from mouse (Sigma-Aldrich) or anti-HA affinity matrix (Roche) for 2–3 hr at 4°C. After incubation, beads with bound protein complexes were washed in ice-cold lysis buffer. Subsequently, 4× NuPAGE sample buffer was added to the beads and heated for 10 min at 70°C. Beads were precipitated by centrifugation, and supernatant was analyzed on NuPAGE Novex 4%–12% Bis-Tris SDS-PAGE gels. After blotting overnight at 4°C, blots were stained with mouse anti-FLAG or mouse anti-HA. Fluorescence was analyzed on a LI-COR Odyssey 2.1 infrared scanner. Full unprocessed western blotting scans are shown in Figure S5. PIH1D3 was consistantly able to precipitate DNAAF2/KTU and DNAAF4/DYX1C1 and vice versa (Figure 4B). Our results using human cells confirm findings by Dong et al.41 for mouse sperm. Similarly to the murine Pih1d3, human PIH1D3 interacts with the chaperone HSP90. In addition we detected protein-protein interactons between PIH1D3 and DNAAF2/KTU as well as DNAAF4/DYX1C1, indicating that PIH1D3 is possibly involved in dynein arm assembly and/or important for facilitating assembly enabled by these other DNAAFs (Figure 4C). Interestingly, in contrast to DNAAF2/KTU or DNAAF4/DYX1C1 deficiency that results in absence of ODA type 2,15, 22 deficiency of PIH1D3 results in absence of ODAs type 1 and type 2. This observation indicates that the assembly of ODA type 1 is PIH1D3 dependent and can occur independently from DNAAF2/KTU or DNAAF4/DYX1C1. Our results show an essential role for human PIH1D3 in the cytoplasmic preassembly process of both ODA types and IDAs, probably by stabilizing the formation of the DNAI1-DNAI2 complex.
Figure 4.
Identification of PIH1D3 Interacting Genes
(A) Y2H assays show strong interaction of PIH1D3 with DNAAF2 and DNAAF4. Lamin-C-pBD- Gal4-pAD was used as a negative control and Gal4-pBD - Gal4-pAD as a positive control.
(B) Confirmation of PIH1D3 interactors from the Y2H screen by co-IP from HEK293T cells. Co-expressions of tagged versions of PIH1D3 and either DNAAF2 or DNAAF4 revealed protein-protein interactions with DNAAF2 and DNAAF4 (left, western-blot anti-Flag; right, western blot anti-HA).
Here we describe mutations in PIH1D3, a gene on the X chromosome, in PCD-affected individuals without coseggregation of severe intellectual disability or retinitis pigmentosa. This is of major clinical importance as symptoms related to primary ciliary dyskinesia were not the primary cause of diagnostic procedures undertaken in individuals described with pathogenic variants in cases presenting with mutations in the X-linked genes RPGR and OFD1.38, 39, 45 Because mutations in other PCD-associated genes usually present with an autosomal-recessive mode of inheritance, it is possible that X-linked inheritance might be overlooked, especially in male cases (1) without syndromic coseggregation, (2) without knowledge of the entire pedigree, or (3) without siblings or with only female siblings. Additionally, loss of function of PIH1D3 results also in sperm immotility and male infertility caused by disrupted assembly of ODAs in sperms, similar to findings observed in DNAAF2/KTU mutant individuals.15 Although it was previously shown that knockout of Pih1D3 in mice results only in sperm defects,17 here we show that human PIH1D3, located on the X chromosome, encodes a dynein axonemal assembly factor in respiratory cilia and sperm flagella. PIH1D3 loss-of-function mutations cause classic PCD phenotype with ciliary and sperm immotility in humans.
Acknowledgments
We are grateful to the affected individuals and their family members whose cooperation made this study possible, and we thank all referring physicians. We thank Prof. J. Neesen (Medical University of Vienna) for providing us anti-DNALI1 antibodies. We thank J. Fritz, Y. Moutaouakil, R.J.P. Musters, J. Kole, D. Ernst, M. Herting, B. Lechtape, L. Overkamp, A. Robbers, F.J. Seesing, and C. Westermann for excellent technical work. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
This work was supported by Fonds NutsOhra (1103-054 to G.P.), the Deutsche Forschungsgemeinschaft grants OM 6/4, OM 6/7, OM 6/8 (H. Omran), the IZKF Muenster to H. Omran (Om2/009/12 and Om/015/16), by the European Commission (FP7/2007–2013) grant agreement (GA) Nr. 262055 (H. Omran) as a Transnational Access project of the European Sequencing and Genotyping Infrastructure (ESGI), and by the European Union seventh FP under GA Nr. 241955, project SYSCILIA (H. Omran), and GA Nr. 305404, project BESTCILIA (H. Omran). M.S. is funded by a Radboudumc Hypatia Tenure track fellowship, a Radboud University Excellence Fellowship, and ERC starting grant TREATCilia (grant agreement 716344) and acknowledges funding from the German Research Foundation (CRC-1140, KIDGEM).
Published: December 29, 2016
Footnotes
Web Resources
- ExAC Browser, http://exac.broadinstitute.org/
- Expression Atlas, http://www.ebi.ac.uk/gxa/home
- GenBank, http://www.ncbi.nlm.nih.gov/genbank/
- OMIM, http://www.omim.org/
- The Human Protein Atlas, http://www.proteinatlas.org/
- varbank, https://varbank.ccg.uni-koeln.de
Supplemental Data
Document S1. Figures S1–S5 and Tables S1–S3
Movie S1. High-Speed Videomicroscopy of PIH1D3 Mutant Respiratory Cilia from OP-1899 II1
Movie S2. High-Speed Videomicroscopy of Normal Respiratory Cilia
Movie S3. High-Speed Videomicroscopy of PIH1D3 Mutant Respiratory Cilia from PCD-10 I1
Movie S4. High-Speed Videomicroscopy of Normal Respiratory Cilia
Movie S5. High-Speed Videomicroscopy of PIH1D3 Mutant Sperm Cell from OP-1899 II1
Movie S6. High-Speed Videomicroscopy of PIH1D3 Normal Sperm Cell
Document S2. Article plus Supplemental Data
References
- 1.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 2.Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Summers K.E., Gibbons I.R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satir P., Heuser T., Sale W.S. A structural basis for how motile cilia beat. Bioscience. 2014;64:1073–1083. doi: 10.1093/biosci/biu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson K.S., Gonzalez O., Dutcher S.K., Bayly P.V. Dynein-deficient flagella respond to increased viscosity with contrasting changes in power and recovery strokes. Cytoskeleton (Hoboken) 2015;72:477–490. doi: 10.1002/cm.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowkes M.E., Mitchell D.R. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell. 1998;9:2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswanadha R., Hunter E.L., Yamamoto R., Wirschell M., Alford L.M., Dutcher S.K., Sale W.S. The ciliary inner dynein arm, I1 dynein, is assembled in the cytoplasm and transported by IFT before axonemal docking. Cytoskeleton (Hoboken) 2014;71:573–586. doi: 10.1002/cm.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowles M.R., Daniels L.A., Davis S.D., Zariwala M.A., Leigh M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison M.J., Shapiro A.J., Kennedy M.P. Congenital heart disease and primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016;18:25–32. doi: 10.1016/j.prrv.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Horani A., Ferkol T., Dutcher S., Brody S. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016;18:18–24. doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibañez-Tallon I., Pagenstecher A., Fliegauf M., Olbrich H., Kispert A., Ketelsen U.-P., North A., Heintz N., Omran H. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum. Mol. Genet. 2004;13:2133–2141. doi: 10.1093/hmg/ddh219. [DOI] [PubMed] [Google Scholar]
- 12.Olbrich H., Häffner K., Kispert A., Völkel A., Volz A., Sasmaz G., Reinhardt R., Hennig S., Lehrach H., Konietzko N. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002;30:143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 13.Pennarun G., Escudier E., Chapelin C., Bridoux A.M., Cacheux V., Roger G., Clément A., Goossens M., Amselem S., Duriez B. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am. J. Hum. Genet. 1999;65:1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panizzi J.R., Becker-Heck A., Castleman V.H., Al-Mutairi D.A., Liu Y., Loges N.T., Pathak N., Austin-Tse C., Sheridan E., Schmidts M. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loges N.T., Olbrich H., Fenske L., Mussaffi H., Horvath J., Fliegauf M., Kuhl H., Baktai G., Peterffy E., Chodhari R. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am. J. Hum. Genet. 2008;83:547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazor M., Alkrinawi S., Chalifa-Caspi V., Manor E., Sheffield V.C., Aviram M., Parvari R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am. J. Hum. Genet. 2011;88:599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duriez B., Duquesnoy P., Escudier E., Bridoux A.-M., Escalier D., Rayet I., Marcos E., Vojtek A.-M., Bercher J.-F., Amselem S. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA. 2007;104:3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartoloni L., Blouin J.-L., Pan Y., Gehrig C., Maiti A.K., Scamuffa N., Rossier C., Jorissen M., Armengot M., Meeks M. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc. Natl. Acad. Sci. USA. 2002;99:10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwabe G.C., Hoffmann K., Loges N.T., Birker D., Rossier C., de Santi M.M., Olbrich H., Fliegauf M., Failly M., Liebers U. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat. 2008;29:289–298. doi: 10.1002/humu.20656. [DOI] [PubMed] [Google Scholar]
- 20.Knowles M.R., Leigh M.W., Carson J.L., Davis S.D., Dell S.D., Ferkol T.W., Olivier K.N., Sagel S.D., Rosenfeld M., Burns K.A., Genetic Disorders of Mucociliary Clearance Consortium Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–441. doi: 10.1136/thoraxjnl-2011-200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onoufriadis A., Paff T., Antony D., Shoemark A., Micha D., Kuyt B., Schmidts M., Petridi S., Dankert-Roelse J.E., Haarman E.G., UK10K Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knowles M.R., Leigh M.W., Ostrowski L.E., Huang L., Carson J.L., Hazucha M.J., Yin W., Berg J.S., Davis S.D., Dell S.D., Genetic Disorders of Mucociliary Clearance Consortium Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hjeij R., Onoufriadis A., Watson C.M., Slagle C.E., Klena N.T., Dougherty G.W., Kurkowiak M., Loges N.T., Diggle C.P., Morante N.F.C., UK10K Consortium CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallmeier J., Shiratori H., Dougherty G.W., Edelbusch C., Hjeij R., Loges N.T., Menchen T., Olbrich H., Pennekamp P., Raidt J. TTC25 deficiency results in defects of the outer dynein arm docking machinery and primary ciliary dyskinesia with left-right body asymmetry randomization. Am. J. Hum. Genet. 2016;99:460–469. doi: 10.1016/j.ajhg.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werner C., Onnebrink J.G., Omran H. Diagnosis and management of primary ciliary dyskinesia. Cilia. 2015;4:2. doi: 10.1186/s13630-014-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loges N.T., Olbrich H., Becker-Heck A., Häffner K., Heer A., Reinhard C., Schmidts M., Kispert A., Zariwala M.A., Leigh M.W. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A.-M., Coste A., Deschildre A., de Blic J., Legendre M., Montantin G. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N.T., Hagiwara H., Zhang Q., Leblond G., O’Toole E., Hara C. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchison H.M., Schmidts M., Loges N.T., Freshour J., Dritsoula A., Hirst R.A., O’Callaghan C., Blau H., Al Dabbagh M., Olbrich H. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012;44:381–389. doi: 10.1038/ng.1106. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarkar A., Loges N.T., Slagle C.E., Francis R., Dougherty G.W., Tamayo J.V., Shook B., Cantino M., Schwartz D., Jahnke C., UK10K DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013;45:995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horani A., Druley T.E., Zariwala M.A., Patel A.C., Levinson B.T., Van Arendonk L.G., Thornton K.C., Giacalone J.C., Albee A.J., Wilson K.S. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kott E., Duquesnoy P., Copin B., Legendre M., Dastot-Le Moal F., Montantin G., Jeanson L., Tamalet A., Papon J.-F., Siffroi J.-P. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore D.J., Onoufriadis A., Shoemark A., Simpson M.A., zur Lage P.I., de Castro S.C., Bartoloni L., Gallone G., Petridi S., Woollard W.J. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;93:346–356. doi: 10.1016/j.ajhg.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zariwala M.A., Gee H.Y., Kurkowiak M., Al-Mutairi D.A., Leigh M.W., Hurd T.W., Hjeij R., Dell S.D., Chaki M., Dougherty G.W. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 2013;93:336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knowles M.R., Ostrowski L.E., Loges N.T., Hurd T., Leigh M.W., Huang L., Wolf W.E., Carson J.L., Hazucha M.J., Yin W. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am. J. Hum. Genet. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austin-Tse C., Halbritter J., Zariwala M.A., Gilberti R.M., Gee H.Y., Hellman N., Pathak N., Liu Y., Panizzi J.R., Patel-King R.S. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budny B., Chen W., Omran H., Fliegauf M., Tzschach A., Wisniewska M., Jensen L.R., Raynaud M., Shoichet S.A., Badura M. A novel X-linked recessive mental retardation syndrome comprising macrocephaly and ciliary dysfunction is allelic to oral-facial-digital type I syndrome. Hum. Genet. 2006;120:171–178. doi: 10.1007/s00439-006-0210-5. [DOI] [PubMed] [Google Scholar]
- 39.Moore A., Escudier E., Roger G., Tamalet A., Pelosse B., Marlin S., Clément A., Geremek M., Delaisi B., Bridoux A.-M. RPGR is mutated in patients with a complex X linked phenotype combining primary ciliary dyskinesia and retinitis pigmentosa. J. Med. Genet. 2006;43:326–333. doi: 10.1136/jmg.2005.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto R., Hirono M., Kamiya R. Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J. Cell Biol. 2010;190:65–71. doi: 10.1083/jcb.201002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong F., Shinohara K., Botilde Y., Nabeshima R., Asai Y., Fukumoto A., Hasegawa T., Matsuo M., Takeda H., Shiratori H. Pih1d3 is required for cytoplasmic preassembly of axonemal dynein in mouse sperm. J. Cell Biol. 2014;204:203–213. doi: 10.1083/jcb.201304076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 43.Dougherty G.W., Loges N.T., Klinkenbusch J.A., Olbrich H., Pennekamp P., Menchen T., Raidt J., Wallmeier J., Werner C., Westermann C. DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am. J. Respir. Cell Mol. Biol. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheway G., Schmidts M., Mans D.A., Szymanska K., Nguyen T.M., Racher H., Phelps I.G., Toedt G., Kennedy J., Wunderlich K.A., UK10K Consortium. University of Washington Center for Mendelian Genomics An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat. Cell Biol. 2015;17:1074–1087. doi: 10.1038/ncb3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bukowy-Bieryłło Z., Ziętkiewicz E., Loges N.T., Wittmer M., Geremek M., Olbrich H., Fliegauf M., Voelkel K., Rutkiewicz E., Rutland J. RPGR mutations might cause reduced orientation of respiratory cilia. Pediatr. Pulmonol. 2013;48:352–363. doi: 10.1002/ppul.22632. [DOI] [PubMed] [Google Scholar]
- 46.Omran H., Loges N.T. Immunofluorescence staining of ciliated respiratory epithelial cells. Methods Cell Biol. 2009;91:123–133. doi: 10.1016/S0091-679X(08)91007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S5 and Tables S1–S3
Movie S1. High-Speed Videomicroscopy of PIH1D3 Mutant Respiratory Cilia from OP-1899 II1
Movie S2. High-Speed Videomicroscopy of Normal Respiratory Cilia
Movie S3. High-Speed Videomicroscopy of PIH1D3 Mutant Respiratory Cilia from PCD-10 I1
Movie S4. High-Speed Videomicroscopy of Normal Respiratory Cilia
Movie S5. High-Speed Videomicroscopy of PIH1D3 Mutant Sperm Cell from OP-1899 II1
Movie S6. High-Speed Videomicroscopy of PIH1D3 Normal Sperm Cell
Document S2. Article plus Supplemental Data