Jumping species—a mechanism for coronavirus persistence and survival (original) (raw)
Abstract
Zoonotic transmission of novel viruses represents a significant threat to global public health and is fueled by globalization, the loss of natural habitats, and exposure to new hosts. For coronaviruses (CoVs), broad diversity exists within bat populations and uniquely positions them to seed future emergence events. In this review, we explore the host and viral dynamics that shape these CoV populations for survival, amplification, and possible emergence in novel hosts.
Highlights
- •
Unique aspects of bat physiology permit and shape the pools of viruses that they harbor. - •
Coronavirus emergence must balance overcoming barriers with maintaining essential viral functions. - •
Spike binding and cleavage is necessary for infection of new species. - •
Coronaviruses acquire tools to disrupt host immunity and maintain viral processes.
Current Opinion in Virology 2017, 23:1–7
This review comes from a themed issue on Viral pathogenesis
Edited by Raul Andino and Michael Diamond
For a complete overview see the Issue and the Editorial
Available online 31st March 2017
Edited by Michael Diamond
http://dx.doi.org/10.1016/j.coviro.2017.01.002
1879-6257/© 2017 The Authors. Published by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
In the past decade, molecular techniques have expanded identification of zoonotic viruses, including coronaviruses (CoVs) [1]. Traditionally, approaches for viral identification have included culturing, antigen staining, electron microscopy, and serology [2]; however, these techniques were inherently biased towards known viral families and were largely insensitive to uncharacterized species. In contrast, molecular diagnostics rapidly identified unknown pathogens starting with Sin Nombre virus in the late 20th century, continuing with SARS-CoV in the early part of this century, and most recently with MERS-CoV [3, 4, 5]. As the molecular approaches improved, these techniques have become standard in identifying infectious agents in both acute and chronic disease settings. Coupled with reduced cost, these new approaches have permitted application for pathogen discovery; the number of known CoVs has increased substantially, aided by both surveys of animal populations and infrastructure investments to improve diagnostic capacity in disease hotspots [6]. Importantly, the resulting inventory illustrates the broad diversity harbored in zoonotic hosts and the presence of quasi-species that may serve as a reservoir for CoV persistence. In this review, we examine how both bat hosts and the CoVs that they harbor may be uniquely positioned to seed future emergence events, especially as human populations increase and penetrate the undeveloped regions of the world.
Bats reservoirs: shaping virus emergence
While numerous animals have been surveyed in the past decade, bats continue to be among the most abundant source for novel viral sequences [7]. Bat species are among the oldest mammals and represent 20% of mammalian diversity [8]; they exist and occupy diverse niches from isolated individuals to large commensal colonies with broad geographic ranges that can span thousands of miles. Importantly, their great diversity and long co-evolutionary relationships with pathogens provide the opportunity for cross species mixing and maintenance of quasi-species pools of viruses that can infect a range of hosts [9, 10]. Yet, despite harboring such a diverse assortment of viruses, surveyed bats rarely exhibit signs of disease. Several hypotheses have been proposed to explain these asymptomatic infections. One postulates that bats, the only flying mammal, produce large amounts of reactive oxygen species (ROS) and, in response, have modulated genes to limit oxidative stress [11], which may result in reduced viral replication and pathogenesis [12]. Similarly, a modified innate immune response may also contribute to the diverse viral pools harbored by bats. Known PYHIN (PYRIN and HIN domain-containing) genes within the inflammasome pathway and natural killer immunoglobulin-like receptors (KIRs) are absent or significantly reduced in some surveyed bat species, potentially limiting disease and damage following infection [11, 13]. In addition, constitutive expression of bat interferon subtypes likely limits disease but permits low-level viral infection to remain intact [14]. A third possibility suggests a commensal relationship between the harbored viruses and bat species [15]. As primarily identified from enteric samples (i.e., bat guano), these pools of viruses may serve a critical role in the bat microbiome to prime immunity, a concept similarly proposed for humans with herpes viruses [16]. Finally, enteric infection represents a significantly different tissue than the respiratory tract in terms of disease and adaptive immunity; thus, virus tropism differences between species and tissues may also contribute to limiting disease in bats. Similarly, while recent work has shown intact elements of adaptive immunity in bat species [17, 18, 19], the enteric location may generate a dampened adaptive response that permits viral maintenance similar to the members of the microbiome in humans [20]. Together, these factors likely work in combination and indicate how diverse pools of CoV quasi-species can survive in bat populations.
While bat species maintain factors that permit virus persistence, the unique host environment also promotes broad diversity in CoV quasi-species pools. As a result of flight, accumulation of ROS species may occur for short periods of time and have been shown to have mutagenic effects, potentially overwhelming CoV proofreading repair and/or altering viral polymerase fidelity and increasing species diversity, a possible key to cross-species transmission [21]. Similarly, the constitutive expression of type I IFN in bat hosts may select for advantageous viral mutations that enhance resistance to innate immune antiviral defense pathways and provide a replication advantage, especially after cross species transmission [14]. Conversely, the absence of key inflammatory mediators in bat species provides no selective pressure to minimize these responses [13]; subsequently, infection of a new host could result in massive and pathogenic inflammation responses, as seen with both SARS-CoV and MERS-CoV infections in humans [22, 23]. Overall, the unique aspects that permit quasi-species pools of viruses in bats also contribute to their diversity and potential to emerge in new species.
Balancing act: honing CoV survival and emergence
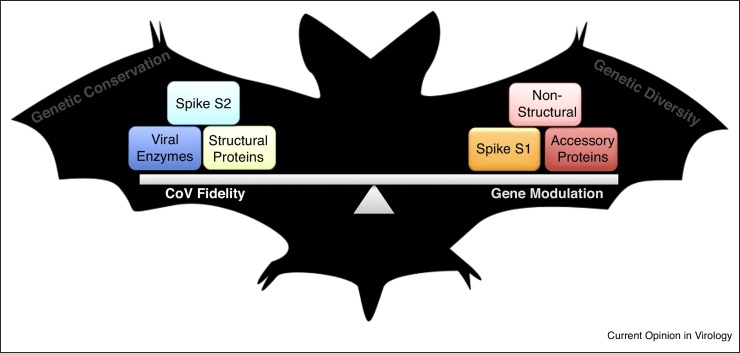
While bats provide a critical foreground, emergence of CoVs requires that key viral factors be altered to overcome species barriers without sacrificing the form or function of other important elements. This dichotomy in CoVs is governed by two distinct mechanisms: fidelity and gene acquisition (Figure 1). A major limitation to RNA virus capacity is the need to minimize sequence length to survive error catastrophe [24]. However, CoVs, as some of the largest members of the Nidovirales order, have overcome this barrier by producing a large replication complex with known RNA synthesis and modification activities that include a proofreading machine, mediated primarily via the 3′–5′ exoribonuclease activity of non-structural protein (nsp) 14 [25]. As such, this large and complex RNA replication machinery has allowed CoVs to achieve upwards of 32 kb in size while maintaining the functional components required for viability. Coupled with robust fidelity, CoVs have also used recombination, horizontal gene transfer, gene duplication, and alternative open reading frames to expand the functional capacity for its current and new hosts [26]. Together, both fidelity and gene acquisition have honed and refined CoV proteins, which can be divided into three broad groups based on selective pressure: spike, conserved, and variable proteins (Figure 1). For a novel CoV to emerge, these three groups must function in harmony, providing sufficient changes to overcome species barriers while maintaining key viral functions.
Figure 1.
Balancing coronavirus emergence. Bat populations maintain a unique environment that facilitates survival and maintenance of diverse pools of viruses. To overcome species barriers, CoV must modify some key viral factors while maintaining others. Two mechanisms govern this balance: fidelity and gene modulation. Using these processes, CoVs shape their proteins conserving some (viral enzymes, structural proteins, spike S2) while modifying others (non-structural proteins, accessory proteins, spike S1). The resulting pools therefore maintain viability while also possessing tools necessary for emergence.
Keying in: spike drives emergence
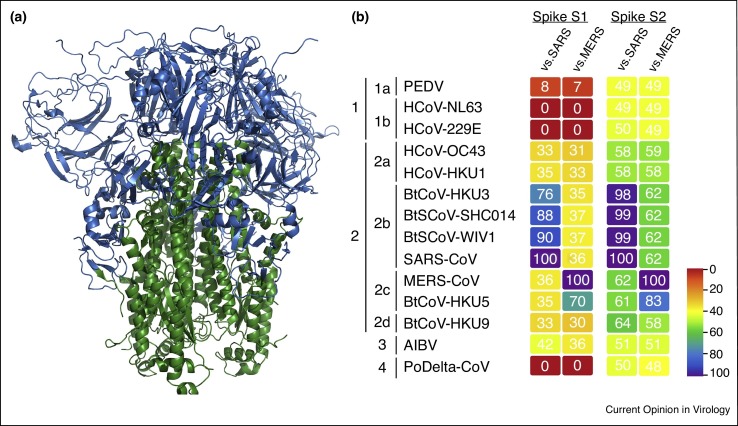
Charged with binding the host receptor, the spike protein of CoVs governs species specificity and is a critical target for host immunity [27]. Divided into two parts, the S1 portion forms the globular head of the spike trimer (Figure 2a), drives receptor engagement, and is variable across and within CoV groups (Figure 2b) [28, 29]. In contrast, the S2 domain maintains the entry machinery and requires more conservation across the CoV family (Figure 2a,b). With binding required for infection, mutations within S1, and most notably, the receptor-binding domain (RBD), have been thought to be critical for CoV emergence [30]. Using chimeric viruses employing civet, early, and middle-phase spike proteins demonstrated viability for the closely related strains in human cells [31, 32]. However, for some strains, such as SZ16 and bat-derived HKU3-CoV, the closest known SARS-CoV progenitors at the time, progeny virions were not recoverable in Vero or primary human airway epithelial cells, despite evidence of RNA replication [30, 32]. To overcome this barrier, single humanizing mutation K479N was introduced into SZ16 and a chimeric HKU3 virus containing the RBD of SARS-CoV was designed and permitted replication, likely due to its capacity to bind the human ACE2 receptor [30, 31]. A similar approach was used with group 2C CoV HKU5; substitution of the entire ecto-domain from SARS-CoV spike resulted in an HKU5 virus that was able to infect human cells [33]. Together, the data argue that the ability of the spike to bind receptor is required for viability in novel hosts.
Figure 2.
Conservation and modification of spike protein. The CoV spike protein is critical receptor binding and entry. Therefore, while modification is likely required for infection of new species, the spike protein must also maintain its entry mechanism. (a) Structure of MHV-CoV spike trimer (adapted from Ref. [53]), dividing the protein into S1 globular head portions (blue), and S2 conserved stalk (green). (b) Heat maps were constructed from a set of representative coronaviruses from all four genogroups using alignment data paired with neighbor-joining phylogenetic trees built in Geneious (v.9.1.5) and visualized in EvolView (evolgenius.info). Trees show the degree of genetic similarity of S1 and S2 domains of the spike glycoprotein.
However, more recent advances identified bat CoV spike proteins that could produce robust infection without manipulation [34, 35]. Building from sequences closely related to the epidemic SARS-CoV strains [36], chimeric viruses employing the spike sequences from SHC014 and WIV1 clusters produced CoVs capable of replicating in human cells and causing disease in vivo [34, 35]. Coupled with the discovery of sequences even more closely related to the epidemic SARS-CoV strains and evidence of robust S1 recombination [37], the results suggest that extensive mutation of the spike RBD may not be the only correlate for infection of human hosts. Notably, both chimeric viruses were attenuated relative to the epidemic strain, suggesting that adaptation within the new host contributes to disease and pathogenesis [34, 35]. Yet, it remains unclear if these mutations occur exclusively within the S1 portion of spike or if subtle changes in the S2 region contribute to enhanced disease by interfacing with surface and intracellular proteases that function in entry and egress [38, 39].
Mainstays and accessories: adding tools but keeping a base
The CoV spike protein captures a critical dichotomy necessary for emergence, employing enough novelty in its S1 region to bind new host receptors while conserving functional entry activity in its S2 portion. However, while critical for infection of new hosts, changing the spike protein alone is not sufficient to cause epidemic disease [34, 35]; therefore, changes within the backbone are also necessary to speed emergence. Yet, the same dichotomy seen with the spike glycoproteins is necessary in balancing change within the CoV backbone. Certain elements, most notably accessory proteins, may be added or modified to enhance infection within new hosts. In contrast, other viral motifs and proteins must be conserved to maintain virus functionality. For each, CoV fidelity, recombination, and evolutionary pressure hone and refine these genes, providing a framework for emergence in a new species to occur.
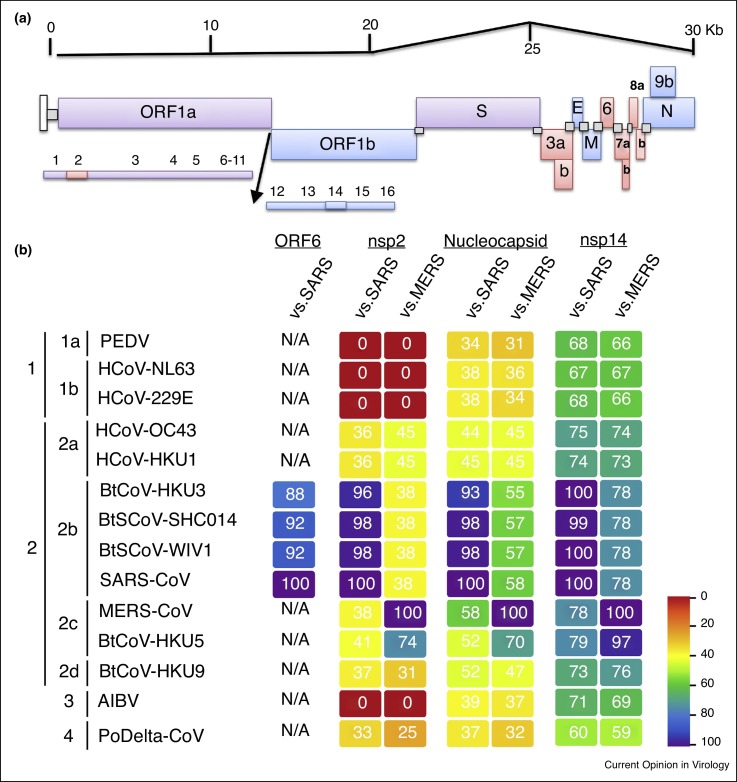
For highly conserved viral functions, the presence of CoV fidelity machinery provides an important means to maintain these activities in the context of an expansive genome. Broadly, these conserved viral proteins can be categorized into structural and enzymatically active groups (Figure 3a). For structural proteins, including the nucleocapsid (N), matrix (M), and envelope (E), high within-group conservation is maintained, with more modest similarity seen across the entire CoV family (Figure 3b). This level of conservation, similar to the S2 portion of spike, suggests the need to maintain functional interaction for the formation of viral particles. Similarly, ORF1ab polyprotein genes find a distinction, with genes involved in protease cleavage and the replication complex having high levels of similarity across CoV families. For example, enzymatically active proteins, such as nsp14 and nsp16, maintain very high conservation, likely due to their specific functions in proofreading and 2′O methylation of nascent RNA [25, 40] (Figure 3). For both groups, some mutational space is available, accounting for differences across the family; however, function must be maintained to ensure CoV survival.
Figure 3.
Maintenance and change the CoV backbone. Changes to the CoV backbone can aid emergence, but must be balanced against conservation of other elements. (a) Genomic structure of SARS-CoV with proteins predicted to be conserved (blue), variable (red), or in between (purple). (b) Heat maps were constructed from a set of representative coronaviruses from all four genogroups using alignment data paired with neighbor-joining phylogenetic trees built in Geneious (v.9.1.5) and visualized in EvolView (evolgenius.info). Trees show the degree of genetic similarity of ORF6, NSP2, nucleocapsid, and NSP14 across genera.
In contrast, accessory proteins distinguish CoV infections from each other, with high variability across the family, allowing viruses to adapt to current and novel hosts. The majority of these genes have been characterized in the context of antagonizing host immune responses, most notably type I IFN pathways [41]. However, the functions of these proteins may extend beyond host immunity and may be species-specific. For example, the SARS-CoV accessory protein ORF6 was initially characterized based on its capacity to interfere with STAT1 nuclear localization [42]. Further study indicated that modulation of the IFN responses was a byproduct of karyopherin transport and had a significant impact on host modulation beyond type I IFN at late times post-infection [42, 43]. Notably, protein-coding sequences similar to SARS-CoV ORF6 are not readily detected beyond the group 2B CoV family, suggesting a more recent acquisition (Figure 3). Similarly, SARS ORF8 has undergone significant modification, with a 29-nucleotide deletion found in epidemic strains resulting in two novel proteins (ORF8a and 8b) [44]; coupled with reports of human isolates with larger deletions, these results suggest that the epidemic strain may be removing a protein only necessary for survival in bats [45]. Even for viral genes within the ORF1ab polyprotein, significant changes can be noted across viral families. Nsp2, cleaved co-translationally from nsp3 and present in some form in all CoV, is responsible for a wide variety of activities and has minimal cross-genus sequence homology, although within groups, similarities are variable (Figure 3) [46, 47, 48]. Together, these results argue that across the CoV family, significant differences in accessory proteins can modulate and change infection aspects, including kinetics, severity, and species.
Yet, even within more closely related subgroups, novel genes can appear from diverse sources and potentially fuel emergence. The recent discovery and characterization of two closely related SARS-like viruses, WIV1 and WIV16, revealed a novel acc.essory protein, ORFX, which was not found in the epidemic SARS-CoV strains [49]. Containing no sequence homology to any known proteins, the novel gene modulates type I IFN and activates NFkB signaling pathways, suggesting a role in modulating host immunity. While the majority of accessory proteins are thought to be acquired from the host, recent work suggests that novel CoV proteins can even be taken from other pathogens [50]. Identification of a novel coronavirus (Ro-BatCoV GCCDC1) also revealed the presence of a unique 3′ protein with homology to a known reovirus gene; a similar finding with the hemaglutinin-esterase in a subset of CoV further suggests the possibility of recombination events occurring between viral families [8, 51]. Together, the results indicate that CoVs can sample, acquire, and maintain a range of diverse proteins that may be critical for maintenance in natural hosts and emergence in new species.
Conclusion
With permissive natural hosts and inherent tools to balance gene modulation/maintenance, CoVs are uniquely positioned to emerge in novel hosts. For both the epidemic strains (SARS and MERS-CoV) and contemporary human strains (HCov 229E, NL63, OC43), significant human disease may be the outcome of cross-species transmission. Importantly, opportunities exist to utilize metagenomics data to prepare and possibly mitigate future emergence events. In seeking these goals, researchers need to consider the factors that drive emergence. In determinations of potential threats, exploring the variable spike S1 portion of bat CoVs to identify viruses capable of binding to human receptors is key. Similarly, targeting highly conserved genes like the S2 region of spike has allowed for the development of therapeutics with broad efficacy against current and potential future CoVs that emerge [28, 52]. In addition, understanding the mechanisms and impact of highly variable genes provides another metric for threat and identifies targets for the generation of attenuated vaccine strains. Together, these approaches provide a platform to leverage our understanding of how CoVs emerge from bat sources to prepare and potentially stem future disease outbreaks. With globalization, habitat loss in developing nations, and uneven public health infrastructures, the survival and amplification of novel CoVs in bat populations is now a lurking threat that requires immediate attention and preparation.
Acknowledgements
Research in this manuscript was supported by grants from the National Institute of Allergy & Infectious Disease and the National Institute of Aging of the NIH under awards U19AI109761, U19AI107810, and AI110700 to RSB; K99AG049092 to VDM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Morse S.S., Mazet J.A., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandl J.N., Ahmed R., Barreiro L.B., Daszak P., Epstein J.H., Virgin H.W., Feinberg M.B. Reservoir host immune responses to emerging zoonotic viruses. Cell. 2015;160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Nichol S.T., Spiropoulou C.F., Morzunov S., Rollin P.E., Ksiazek T.G., Feldmann H., Sanchez A., Childs J., Zaki S., Peters C.J. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Kreuder Johnson C., Hitchens P.L., Smiley Evans T., Goldstein T., Thomas K., Clements A., Joly D.O., Wolfe N.D., Daszak P., Karesh W.B. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci Rep. 2015;5:14830. doi: 10.1038/srep14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony S.J., Epstein J.H., Murray K.A., Navarrete-Macias I., Zambrana-Torrelio C.M., Solovyov A., Ojeda-Flores R., Arrigo N.C., Islam A., Ali Khan S. A strategy to estimate unknown viral diversity in mammals. MBio. 2013;4 doi: 10.1128/mBio.00598-13. e00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X.M., Kousoulas K.G., Storz J. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology. 1992;186:318–323. doi: 10.1016/0042-6822(92)90089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luis A.D., Hayman D.T., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reshi M.L., Su Y.C., Hong J.R. RNA viruses ROS-mediated cell death. Int J Cell Biol. 2014;2014:467452. doi: 10.1155/2014/467452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn M., Cui J., Irving A.T., Wang L.F. Unique loss of the PYHIN gene family in bats amongst mammals: implications for inflammasome sensing. Sci Rep. 2016;6:21722. doi: 10.1038/srep21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou P., Tachedjian M., Wynne J.W., Boyd V., Cui J., Smith I., Cowled C., Ng J.H., Mok L., Michalski W.P. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc Natl Acad Sci U S A. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynne J.W., Wang L.F. Bats and viruses: friend or foe? PLoS Pathog. 2013;9:e1003651. doi: 10.1371/journal.ppat.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton E.S., White D.W., Cathelyn J.S., Brett-McClellan K.A., Engle M., Diamond M.S., Miller V.L., Virgin H.W. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 17.Wynne J.W., Woon A.P., Dudek N.L., Croft N.P., Ng J.H., Baker M.L., Wang L.F., Purcell A.W. Characterization of the antigen processing machinery and endogenous peptide presentation of a bat MHC class I molecule. J Immunol. 2016;196:4468–4476. doi: 10.4049/jimmunol.1502062. [DOI] [PubMed] [Google Scholar]
- 18.Martinez Gomez J.M., Periasamy P., Dutertre C.A., Irving A.T., Ng J.H., Crameri G., Baker M.L., Ginhoux F., Wang L.F., Alonso S. Phenotypic and functional characterization of the major lymphocyte populations in the fruit-eating bat Pteropus alecto. Sci Rep. 2016;6:37796. doi: 10.1038/srep37796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medeiros R., Jusot V., Houillon G., Rasuli A., Martorelli L., Kataoka A.P., Mechlia M.B., Le Guern A.S., Rodrigues L., Assef R. Persistence of rabies virus-neutralizing antibodies after vaccination of rural population following vampire bat rabies outbreak in Brazil. PLoS Negl Trop Dis. 2016;10:e0004920. doi: 10.1371/journal.pntd.0004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapp K., Maul J., Hostmann A., Mundt P., Preiss J.C., Wenzel A., Thiel A., Zeitz M., Ullrich R., Duchmann R. Modulation of systemic antigen-specific immune responses by oral antigen in humans. Eur J Immunol. 2010;40:3128–3137. doi: 10.1002/eji.201040701. [DOI] [PubMed] [Google Scholar]
- 21.Seronello S., Montanez J., Presleigh K., Barlow M., Park S.B., Choi J. Ethanol and reactive species increase basal sequence heterogeneity of hepatitis C virus and produce variants with reduced susceptibility to antivirals. PLoS One. 2011;6:e27436. doi: 10.1371/journal.pone.0027436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J., Zhao J., Van Rooijen N., Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crotty S., Cameron C.E., Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peck K.M., Burch C.L., Heise M.T., Baric R.S. Coronavirus host range expansion and middle east respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu Rev Virol. 2015;2:95–117. doi: 10.1146/annurev-virology-100114-055029. [DOI] [PubMed] [Google Scholar]
- 27.Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan T., Rockx B., Donaldson E., Corti D., Baric R. Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus. J Virol. 2008;82:8721–8732. doi: 10.1128/JVI.00818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheahan T., Rockx B., Donaldson E., Sims A., Pickles R., Corti D., Baric R. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J Virol. 2008;82:2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agnihothram S., Yount B.L., Jr., Donaldson E.F., Huynh J., Menachery V.D., Gralinski L.E., Graham R.L., Becker M.M., Tomar S., Scobey T.D. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. MBio. 2014;5:e00047–00014. doi: 10.1128/mBio.00047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X.L., Hu B., Wang B., Wang M.N., Zhang Q., Zhang W., Wu L.J., Ge X.Y., Zhang Y.Z., Daszak P. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol. 2016;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y., Liu C., Du L., Jiang S., Shi Z., Baric R.S., Li F. Two mutations were critical for bat-to-human transmission of middle east respiratory syndrome coronavirus. J Virol. 2015;89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menachery V.D., Debbink K., Baric R.S. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims A.C., Tilton S.C., Menachery V.D., Gralinski L.E., Schafer A., Matzke M.M., Webb-Robertson B.J., Chang J., Luna M.L., Long C.E. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol. 2013;87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau S.K., Feng Y., Chen H., Luk H.K., Yang W.H., Li K.S., Zhang Y.Z., Huang Y., Song Z.Z., Chow W.N. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham R.L., Sims A.C., Baric R.S., Denison M.R. The nsp2 proteins of mouse hepatitis virus and SARS coronavirus are dispensable for viral replication. Adv Exp Med Biol. 2006;581:67–72. doi: 10.1007/978-0-387-33012-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cornillez-Ty C.T., Liao L., Yates J.R., 3rd, Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gadlage M.J., Graham R.L., Denison M.R. Murine coronaviruses encoding nsp2 at different genomic loci have altered replication, protein expression, and localization. J Virol. 2008;82:11964–11969. doi: 10.1128/JVI.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng L.P., Gao Y.T., Ge X.Y., Zhang Q., Peng C., Yang X.L., Tan B., Chen J., Chmura A.A., Daszak P. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, involved in modulation of the host immune response. J Virol. 2016;90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang C., Liu W.J., Xu W., Jin T., Zhao Y., Song J., Shi Y., Ji W., Jia H., Zhou Y. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. 2016;12:e1005883. doi: 10.1371/journal.ppat.1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klausegger A., Strobl B., Regl G., Kaser A., Luytjes W., Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F., Du L., Yu F. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]