Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes (original) (raw)
. Author manuscript; available in PMC: 2018 Apr 13.
Published in final edited form as: Science. 2008 Aug 15;321(5891):960–964. doi: 10.1126/science.1159689
Abstract
Prokaryotes acquire virus resistance by integrating short fragments of viral nucleic acid into clusters of regularly interspaced short palindromic repeats (CRISPRs). Here we show how virus-derived sequences contained in CRISPRs are used by CRISPR-associated (Cas) proteins from the host to mediate an antiviral response that counteracts infection. After transcription of the CRISPR, a complex of Cas proteins termed Cascade cleaves a CRISPR RNA precursor in each repeat and retains the cleavage products containing the virus-derived sequence. Assisted by the helicase Cas3, these mature CRISPR RNAs then serve as small guide RNAs that enable Cascade to interfere with virus proliferation. Our results demonstrate that the formation of mature guide RNAs by the CRISPR RNA endonuclease subunit of Cascade is a mechanistic requirement for antiviral defense.
The clusters of regularly interspaced short palindromic repeat (CRISPR)–based defense system protects many bacteria and archaea against invading conjugative plasmids, transposable elements, and viruses (1–8). Resistance is acquired by incorporating short stretches of invading DNA sequences in genomic CRISPR loci (1, 9, 10). These integrated sequences are thought to function as a genetic memory that prevents the host from being infected by viruses containing this recognition sequence. A number of CRISPR-associated (cas) genes (11 –13) has been reported to be essential for the phage-resistant phenotype (1). However, the molecular mechanism of this adaptive and inheritable defense system in prokaryotes has remained unknown.
The Escherichia coli K12 CRISPR/cas system comprises eight cas genes: cas3 (predicted HD-nuclease fused to a DEAD-box helicase), five genes designated casABCDE, cas1 (predicted integrase) (13), and the endoribonuclease gene cas2 (14) (Fig. 1A and table S1). In separate experiments, each Cas protein was tagged at both the N and C terminus and produced along with the complete set of untagged Cas proteins (15). Affinity purification of the tagged component enabled the identification of a protein complex composed of five Cas proteins: CasA, CasB, CasC, CasD, and CasE (Fig. 1B). The complex, denoted Cascade (CRISPR-associated complex for antiviral defense), could be isolated from E. coli lysates using any of the tagged subunits of the complex as bait, except for CasA.
Fig. 1.

The composition of the Cascade complex. (A) Schematic diagram of the CRISPR/cas gene cluster of E. coli K12 W3110. Repeats and spacers are indicated by diamonds and rectangles, respectively. A palindrome in the repeat is marked by convergently pointing arrows. Protein family nomenclature is as described in (11, 12). (B) Coomassie blue—stained SDS-polyacrylamide gel of the affinity purified protein complex using either the N-terminal StrepII-tag (S) or C-terminal His-tag (H) of each of the subunits CasB, CasC, CasD, or CasE as bait. Asterisks indicate the 5.5 kD larger double-tagged subunits. Marker sizes in kilodaltons on the left; location of untagged subunits on the right.
The function of Cascade was studied by analyzing the effect of in-frame cas gene knockouts (16) on the formation of transcripts of the CRISPR region in E. coli K12 (Fig. 1A). Northern analysis of total RNA with single-stranded spacer sequences as a probe showed transcription of the CRISPR region in the direction downstream of the cas2 gene (Figs. 1A and 2A) and no transcription in the opposite direction. Analysis of control strains (wild type and a non-cas gene knockout) revealed a small CRISPR-RNA (crRNA) product of ∼57 nucleotides (Fig. 2A). The same product was present in much higher amounts in the casA, casB, and casC knockout strains but absent from strains lacking the overlapping genes casD and casE (Fig. 2A). The small crRNAs seem to be cleaved from a multi-unit crRNA precursor (pre-crRNA) (7, 17, 18), as is evident from the presence of two and three repeat-spacer units (∼120 and ∼180 nucleotides) that show up in the Δ_casA_, Δ_casB_, and Δ_casC_ strains (Fig. 2A). The Δ_casE_ strain contained a large pre-crRNA, suggesting that the disruption of this gene prevents pre-crRNA cleavage.
Fig. 2.
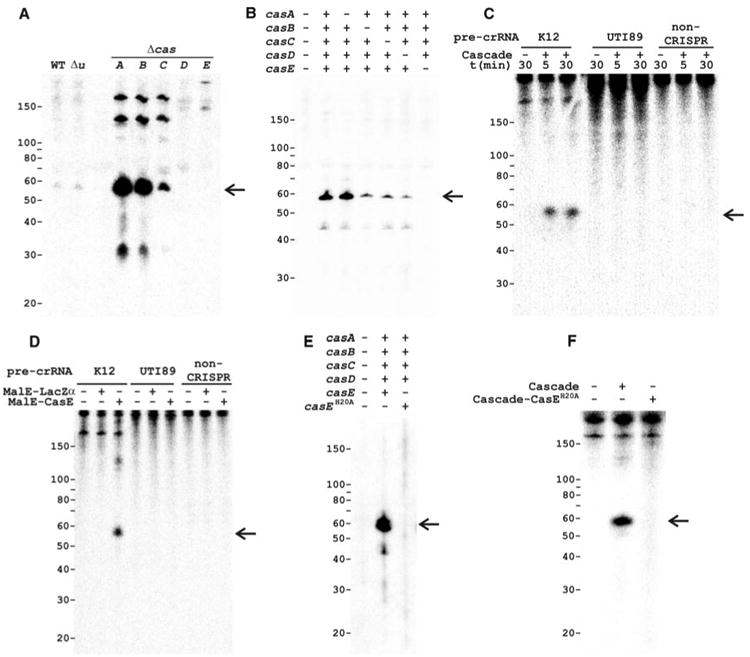
Cascade cleaves CRISPR RNA precursors into small RNAs of ∼57 nucleotides (marked by arrows). (A) Northern analysis of total RNA of WT E. coli K12 (WT), a non-cas gene knockout (Δ_u, uidA_, β-glucuronidase), and Cascade gene knockouts using the single-stranded spacer sequence BG2349 (table S2) as a probe. (B) Northern blot as in (A) of total RNA from E. coli BL21 (DE3) expressing the E. coli K12 pre-crRNA and either the complete or incomplete Cascade complex. (C) Activity assays with purified Cascade using in vitro transcribed α-32P–uridine triphosphate–labeled pre-crRNA from E. coli K12 (repeat sequence: GAGUUCCCCGCCAGCGGGGAUAAACCG), E. coli UTI89 (repeat sequence: GUUCACUGCCGUACAGGCAGCUUAGAAA), and non-crRNA as substrates. (D) Activity assays as shown in (C) for 15 min with purified MalE-LacZα and MalE-CasE fusion proteins. (E) Northern blot as shown in (B) with Cascade or Cascade-CasEH20A. (F) Activity assays as shown in (C) for 30 min with purified Cascade or Cascade-CasEH20A.
To study the accumulation and cleavage patterns of crRNAs in the E. coli K12 knockout strains in more detail and to rule out any effects of the gene disruptions on the expression of downstream or upstream cas genes, the five subunits of Cascade and the K12-type pre-crRNA were expressed in E. coli BL21(DE3), which lacks endogenous cas genes (19). Northern analysis showed that crRNAs of ∼57 nucleotides were only produced in strains containing the Cascade complex (Fig. 2B). By omitting the individual subunits one by one, it became apparent that the small crRNA was absent only in the strain that lacked casE (Fig. 2B), indicating that this is the only Cascade subunit essential for pre-crRNA cleavage.
Activity assays with purified Cascade showed that the complex is capable of cleaving the E. coli K12 pre-crRNA into fragments of ∼57 nucleotides in vitro (Fig. 2C). However, no cleavage was observed with either pre-crRNA from E. coli UTI89, which contains repeats with a different sequence (20), or a non-crRNA template (Fig. 2C). The RNA cleavage reaction proceeded in the absence of divalent metal ions and adenosine triphosphate and reached saturation level within 5 min. To investigate whether the CasE subunit is sufficient for pre-crRNA cleavage activity, it was overproduced as a fusion with the E. coli maltose binding protein (MalE). Like the complete Cascade, the CasE fusion protein cleaved only the K12-type pre-crRNA (Fig. 2D), showing that CasE is an unusual endoribonuclease that does not require the other Cascade subunits. We cannot rule out the possibility that pre-crRNA cleavage is an auto-catalytic, ribozyme-like reaction, in which CasE is an essential RNA chaperone.
CasE belongs to one of the numerous families of repeat-associated mysterious proteins, the largest and most diverse class of Cas proteins (12, 13). The crystal structure of a CasE homolog from Thermus thermophilus HB8 shows that the protein contains two domains with a ferredoxin-like fold, and displays overall structural similarity to a variety of RNA-binding proteins (13, 21). On the basis of structure and amino acid conservation analysis of this protein family (fig. S1), the invariant residue His20 was mutated to Ala to analyze the effect on pre-crRNA cleavage. Northern blots indicated that crRNAs of ∼57 nucleotides were no longer formed in the strain containing Cascade-CasEH20A (Fig. 2E). Moreover, although the mutated CasE was still incorporated into Cascade, the pre-crRNA cleaving ability of purified Cascade was abolished (Fig. 2F), providing further support for the essential role of CasE in pre-crRNA cleavage and suggesting that the conserved His residue is involved in catalysis.
The crRNA cleavage sites were examined by simultaneous expression of K12-type pre-crRNA and Cascade. Under these conditions, the purification of Cascade yielded substantial amounts of copurified RNAs of ∼57 nucleotides (Fig. 3A). Cloning and sequencing of this Cascade-bound RNA revealed that 85% of the clones [67 out of 79 clones (67/79)] were derived from crRNAs, of which 78% (52/67) started with the last eight bases of the repeat sequence (AUAAACCG) (Fig. 3B and fig. S2). This well-defined 5′ end was followed by a complete spacer sequence and a less well-defined 3′ sequence ending in the next repeat region. A transcript of a single palindromic repeat can fold as a stable stem-loop of seven base pairs, which may facilitate recognition by RNA-binding Cas proteins (8, 20), such as CasE. The pre-crRNA cleavage site (PCS) appeared to be located immediately upstream of the 3′ terminal base of the stem-loop formed by the repeat (Fig. 3B). The clone library did not contain crRNAs of 61 nucleotides, which would be the result of a single endonuclease cleavage event in each repeat, given the size of a repeat (29 nucleotides) and most spacers (32 nucleotides). Instead, in agreement with experimental observations (Figs. 2 and 3A), the crRNAs were truncated at the 3′ end by at least two guanosine bases from the endonuclease cleavage site, removing several stem-forming bases.
Fig. 3.
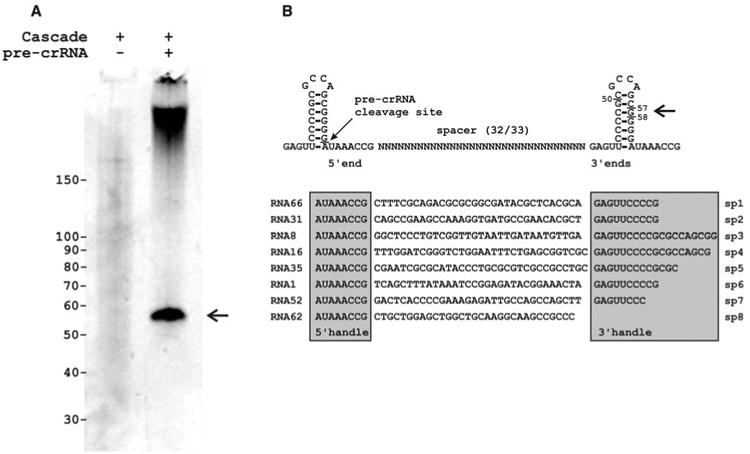
Cleaved crRNAs remain bound by Cascade. (A) Denaturing polyacryl-amide gel showing the crRNA (marked by the arrow) isolated from purified Cascade in the absence and presence of co-expressed pre-crRNA. (B) Secondary structure of pre-crRNA repeats and example sequences of cloned crRNAs indicating the PCS and crRNA handles.
To test whether crRNA-loaded Cascade gives rise to phage resistance, two artificial CRISPRs were designed against phage Lambda (λ). Each of these CRISPRs targeted four essential λ genes (fig. S3). The coding CRISPR (C1–4) produced crRNAs complementary to both the mRNA and the coding strand of these four genes, whereas the template CRISPR (T1–4) targeted only the template strand of the same proto-spacer regions (fig. S3). A nontargeting CRISPR containing wild-type (WT) spacers with no similarity to the phage genome served as a control. Plaque assays with E. coli showed that the introduction of either one of these anti-λ phage CRISPRs in a strain expressing only Cascade did not result in reduced sensitivity of the host to a virulent Lambda phage (λvir) (Fig. 4A). However, strains that expressed Cascade and Cas3 were much less sensitive to phage infection. The template CRISPR rendered the strain insensitive to the phage at the highest phage titer tested (>107-fold less sensitive than the control strain), whereas the coding CRISPR reduced the sensitivity 102fold (Fig. 4A) and produced plaques with a diameter ~1/10 of the standard λ plaque. The phage resistance phenotype was lost when Cascade was omitted (Fig. 4A), proving that both Cascade and Cas3 are required in this process. Moreover, strains containing Cas3 and Cascade-CasEH20A displayed a sensitive phenotype, which shows that pre-crRNA cleavage is mechanistically required for phage resistance. The co-expression of Cas1 and Cas2 had no effect on the sensitivity profile of the strain (Fig. 4A), suggesting that these proteins are involved in other stages of the CRISPR/cas mechanism. Plaque assays with single anti-λ spacers (fig. S3) showed that the total reduction of sensitivity observed with the four anti-λ spacers (C1–4 and T1–4) (Fig. 4A) results from a synergistic effect of the individual spacers (C1 to T4) (Fig. 4B).
Fig. 4.

Engineered CRISPRs confer resistance to λ in the presence of Cascade and Cas3. (A) Effect of the presence of different sets of cas genes on the sensitivity of E. coli to phage λvir. Cells were equipped with one of two engineered CRISPRs containing four anti-λ spacers each (fig. S3). The C1–4 CRISPR produces crRNA complementary to the coding strand and mRNA of λvir, and the T1–4 CRISPR targets only the template strand. The sensitivity of each strain to phage λvir is represented as a histogram of the efficiency of plaquing, which is the plaque count ratio of the anti-λ CRISPR to that of the nontargeting control CRISPR. (B) Effect of single anti-λ spacers (fig. S3) on the sensitivity of E. coli to λvir. Error bars indicate 1 SD.
Our results demonstrate that a complex of five Cas proteins is responsible for the maturation of pre-crRNA to small crRNAs that are critical for mediating an antiviral response. These mature crRNAs contain the antiviral spacer unit flanked by short RNA sequences derived from the repeat on either side termed the 5′ and 3′ handle, which may serve as conserved binding sites for Cascade subunits, as has been suggested previously (20). The Cascade-bound crRNA serves as a guide to direct the complex to viral nucleic acids to mediate an antiviral response. We hypothesize that crRNAs target virus DNA, because anti-λ CRISPRs of both polarities lead to a reduction of sensitivity to the phage. The model is supported by previous observations that virus-derived sequences are integrated into CRISPR loci, irrespective of their orientation in the virus genome (1–4, 7, 9,10,13). We conclude that the transcription of CRISPR regions—and the cleavage of pre-crRNA to mature crRNAs by Cas proteins—is the molecular basis of the antiviral defense stage of the CRISPR/cas system, which enables prokaryotes to effectively prevent phage predation.
Supplementary Material
Supplemental Information
Acknowledgments
We thank T. Verweij, C. G. J. van Houte, and M. R. Beijer for experimental contributions and T. Goosen (Hogeschool van Arnhem en Nijmegen BioCentre), M. J. Young (Montana State University), T. Bisseling, and W. M. de Vos (Wageningen University) for helpful discussions. We are grateful for receiving strains from the KEIO collection distributed by National BioResource Project (National Institute of Genetics, Japan). We thank U. Dobrindt (University of Würzburg) for sending genomic material of E. coli UTI89. This work was financially supported by a Vici grant from the Dutch Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek) and a Marie Curie grant from the European Union. M.L. was supported by the Wenner-Gren Foundations.
Footnotes
References and Notes
- 1.Barrangou R, et al. Science. 2007;315:1709. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. J Mol Evol. 2005;60:174. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 3.Pourcel C, Salvignol G, Vergnaud G. Microbiology. 2005;151:653. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Microbiology. 2005;151:2551. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 5.Godde JS, Bickerton A. J Mol Evol. 2006;62:718. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 6.Tyson GW, Banfield JF. Environ Microbiol. 2008;10:200. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 7.Lillestøl RK, Redder P, Garrett RA, Brügger K. Archaea. 2006;2:59. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorek R, Kunin V, Hugenholtz P. Nat Rev Microbiol. 2008;6:181. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 9.Horvath P, et al. J Bacteriol. 2008;190:1401. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deveau H, et al. J Bacteriol. 2008;190:1390. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen R, Embden JD, Gaastra W, Schouls LM. Mol Microbiol. 2002;43:1565. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 12.Haft DH, Selengut J, Mongodin EF, Nelson KE. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beloglazova N, et al. J Biol Chem. 2008;283:20361. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supporting material on Science Online.
- 16.Baba T, Mori H. Methods Mol Biol. 2008;416:171. doi: 10.1007/978-1-59745-321-9_11. [DOI] [PubMed] [Google Scholar]
- 17.Tang TH, et al. Proc Natl Acad Sci USA. 2002;99:7536. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang TH, et al. Mol Microbiol. 2005;55:469. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim JF, Jeong H, Lenski RE. personal communication [Google Scholar]
- 20.Kunin V, Sorek R, Hugenholtz P. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebihara A, et al. Protein Sci. 2006;15:1494. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information