Cytoplasmic microtubule organization in fission yeast (original) (raw)
. Author manuscript; available in PMC: 2018 May 9.
Published in final edited form as: Yeast. 2006 Oct 15;23(13):1001–1014. doi: 10.1002/yea.1404
Abstract
During the cell cycle of the fission yeast Schizosaccharomyces pombe, striking changes in the organization of the cytoplasmic microtubule cytoskeleton take place. These may serve as a model for understanding the different modes of microtubule organization that are often characteristic of differentiated higher eukaryotic cells. In the last few years, considerable progress has been made in our understanding of the organization and behavior of fission yeast cytoplasmic microtubules, not only in the identification of the genes and proteins involved but also in the physiological analysis of function using fluorescently-tagged proteins in vivo. In this review we discuss the state of our knowledge in three areas: microtubule nucleation, regulation of microtubule dynamics and the organization and polarity of microtubule bundles. Advances in these areas provide a solid framework for a more detailed understanding of cytoplasmic microtubule organization.
Introduction
Microtubules (MTs) are hollow, cylindrical polymers that are found in all eukaryotic cells and are formed by the non-covalent association of tubulin protein molecules [2]. They can assume a variety of distributions in cells and are important for many different large-scale cellular functions, notably cell division and cell polarity, serving alternatively as structural components of major subcellular assemblies and/or as tracks for motor-driven transport of subcellular components. Because of the nature of their assembly from α,β-tubulin dimer, MTs are polar structures, with two distinct ends. These have been designated “plus” and minus” ends, based on polymerization kinetics in vitro [6]; in vivo, the classical picture of MT polarity is one in which MT minus-ends are anchored in MT nucleating sites, while MT plus-ends are able to add or lose additional tubulin dimer subunits and thus grow or shrink, either stochastically or in a regulated manner, depending on their context. In vivo and in vitro, MT plus-ends can often undergo repeated rounds of growing and shrinking, a mechanistically complex process referred to as dynamic instability [18,56].
While the standard picture of MT organization in higher eukaryotic cells is one in which MTs radiate from a perinuclear centrosome, variations on this mode of organization are observed in many types of differentiated cells, such as neurons, muscle cells, or epithelial cells [42]. Relatively little is known about the molecular mechanisms underlying such variant non-centrosomal organization. In the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, the nucleus-associated spindle pole body (SPB) functions as the equivalent of the centrosome. In budding yeast, the SPB is embedded within the nuclear envelope and nucleates both intranuclear mitotic spindle MTs from its nucleoplasmic face and cytoplasmic MTs from its cytoplasmic face. In fission yeast, the situation is more complex. Both intranuclear mitotic spindle MTs and cytoplasmic MTs are nucleated by the fission yeast SPB, but additional sites of cytoplasmic MT nucleation also exist; the distribution of these sites is dynamic and changes during the cell cycle (see below). The difference in cytoplasmic MT organization between budding yeast and fission yeast is also reflected in the relative importance of MTs in cell polarity in the two yeasts; in fission yeast, MTs play a critical role in establishing positions of sites of polarized growth [71,77,78], while in budding yeast they play no such role [38,39].
Cytoplasmic MT organization in fission yeast, with it perhaps tens of MTs, organized in a small number of MT bundles (see below), can thus be regarded as being intermediate between the more simple MT organization of budding yeast and the much more complex organization possible in higher eukaryotic cells, which may have several hundreds of individual MTs. Accordingly, understanding MT organization in fission yeast may provide useful insights into the mechanisms by which complex patterns of MT organization are achieved in higher eukaryotes, especially in differentiated cells. The last 3-5 years have seen substantial progress in our understanding of the molecules and mechanisms controlling fission yeast cytoplasmic MT organization. In this short review, we will focus on three areas: microtubule nucleation, the regulation of microtubule dynamics and the organization and polarity of microtubule bundles. We will not address the assembly or function of MTs in the the intranuclear mitotic spindle, a complex area in its own right.
Nucleating Microtubules
Nucleation sites
During the vegetative (i.e., non-meiotic) cell cycle in fission yeast, three different modes of microtubule organization are present, in succession, nucleated by three different types of microtubule-organizing centers (MTOCs; Fig. 1; see [32] for early references**)**. During interphase, MTs can be nucleated not only from the spindle pole body (SPB) but also from additional sites on the nuclear surface, on microtubules themselves, and in the cytoplasm (see, for example, [20,40,76,89]). These non-SPB sites are generally known collectively as interphase MTOCs (iMTOCs; see below). During mitosis, the cytoplasmic face of the SPB nucleates astral MTs. At the end of mitosis, MTs are nucleated from an equatorial MTOC (eMTOC) at the cell division site (the septum), forming a transient structure, the post-anaphase array (PAA). Neither iMTOCs nor eMTOCs are present in budding yeast Saccharomyces cerevisiae. We will first address more phenomenological aspects of MT nucleation and then discuss the molecules involved.
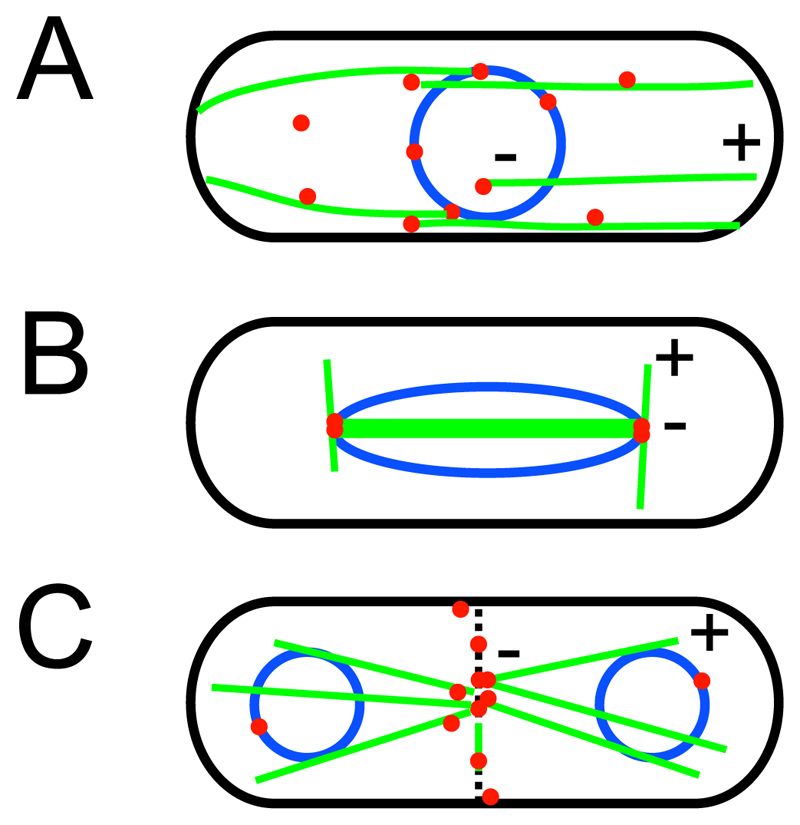
Figure 1. Microtubule organization in the fission yeast cell cycle.
A highly schematic illustration of microtubule (MT) distribution (green) in relation to microtubule organizing centers (MTOCs; red) and the nuclear envelope (blue). During interphase (A), MTOCs may be associated with the nuclear envelope or with existing MTs and may occasionally also be found free in the cytoplasm. MT minus ends (“-“) are generally found towards the cell center and MT plus ends (“+”) towards cell tips. During mitosis (B), intranuclear MTs form the mitotic spindle and astral MTs are nucleated from the SPBs. At the close of mitosis, during cell division (C), the equatoral MTOCs forms at the division site, to nucleate post-anaphase array MTs.
What constitutes an iMTOC?
Ideally, iMTOCs would be defined straightforwardly as the sites from which interphase MTs are normally nucleated. While this definition would not seem controversial, in reality the practical details of experiments have influenced what different workers have termed to be “iMTOCs”, leading to potentially different mechanistic interpretations about the nucleation process itself. In early experiments examining MT regrowth after cold-shock (which depolymerizes MTs much more effectively than MT-destabilizing drugs; see, for example, [76,78]), MTs were found to renucleate predominantly from the nuclear surface [55], and the general conclusion at the time was that this and/or the SPB would be the primary site of iMTOCs [32]. However, characterization of the MT-nucleation protein mto1p (see below) showed that the high degree of MT renucleation from the nuclear surface seen during recovery from cold-shock is due in part to a cold-induced redistribution of mto1p to the nuclear surface from other sites, such as MTs themselves, where mto1p is normally relatively abundant during steady-state growth [76]. Without knowing this, the relative contribution of nuclear envelope-associated iMTOCs to total MT nucleation during normal cell growth could be misjudged.
To assay cytoplasmic interphase MT nucleation from MTs themselves is not possible after a perturbation such as cold-shock, because all MTs are initially depolymerized. Rather, assays of MT nucleation at steady-state require live cell imaging and GFP-tubulin, which, although widely used, may be found to introduce its own subtle artifacts when subject to scrutiny [75,76]. In addition, it is difficult to observe true nucleation of new MTs from existing MTs [40], because of the near-superposition of “old” and “new” signals of GFP-tubulin polymer, and thus we still don’t know how many “iMTOCs” are actually present on existing MTs. To complicate the issue, there is good evidence that MTs can regrow from the overlap region of antiparallel MT bundles in the middle of the cell (see section C, below) [10], but is this true nucleation? These extra-stable MT overlap regions, which can be seen as MT “stubs” after drug-induced MT depolymerization, have been referred to in the literature as iMTOCs (see, for example [10,40]), but it seems likely that the mechanism of MT regrowth from such sites is not the same as true MT nucleation de novo, such as appears to occur on the nuclear envelope or occasionally in the cytoplasm, free of any nearby MT polymer (see, for example, [76]). Finally, other work may refer to “satellites” of γ-tubulin complex proteins as “iMTOCs”, or imply as much (see below), even though it has only recently been possible to observe MTs being nucleated on existing MTs with any confidence [40], and it is indeed possible that only a fraction of these cytologically-defined “satellites” are actually competent for MT nucleation.
In spite of the absence of a universally accepted definition of iMTOCs, the different operational definitions of iMTOCs now in currency all have their value. Rather than try to establish a hard definition for iMTOCs here, we would emphasize that the iMTOC concept is still a loose one, reflecting recent rapid progress in the field as well as the diversity of experimental approaches taken. It is likely that as our understanding improves over time, any differences of opinion over “what is an iMTOC?” will disappear. However, even with a loose definition there are still very focussed questions that can be asked. For example, what are the relative contributions of nuclear-envelope-associated iMTOCs vs. MT-associated iMTOCs to total MT nucleation under steady-state conditions? Are all iMTOCs identical at the molecular level? What localizes them to discrete sites, and by what mechanism are iMTOCs associated with MTs? A particularly mysterious question is what links iMTOCs to the nuclear surface. Possible candidates of interest include the nuclear rim protein amo1p and the TACC homolog mia1/alp7p [68,100].
When do astral MTs appear and what do they do?
It is currently controversial whether cytoplasmic astral MTs exist prior to anaphase. Although there is a literature arguing that they exist and that they are important for a “spindle-orientation checkpoint” (SOC) [27,28,65,72], more recent work suggests that cytoplasmic astral MTs arise only after the metaphase-anaphase transition, making them of dubious value in promoting progression into anaphase. What have been thought to be pre-anaphase cytoplasmic astral MTs may in fact be intranuclear MTs, nucleated from the nucleoplasmic face of the SPB [104], but this issue may still not be resolved to everyone’s satisfaction (see [26] for a more complete overview). While there is not space to debate the merits of the SOC concept here, at this point it seems clear that the concept probably requires at least some revision and/or a critical reassessment, as cells without astral MTs have been observed to progress quickly into anaphase [76,103] (but see also [93]).
If the SOC concept does turn out to be less useful than initially envisioned, the question still remains as to what function astral MTs actually serve. Although it is plausible that astral MTs do play an ancillary role in alignment of the spindle with respect to the cell axis, several types of experiments indicate that they are not required for anaphase B mitotic spindle elongation [53,76,88,93,103]. It is also possible that astral MTs play no significant role at all in mitosis, and that they appear only epiphenomenally, for example because MT nucleation complexes happen to be at the SPB at this stage in the cell cycle.
The eMTOC and the PAA
The eMTOC has not been intensively studied, but what work has been done indicates that its formation involves multiple degrees of control. At a regulatory level, eMTOC formation requires both polo kinase plo1p and components of the multi-gene Septation Initiation Network [35,45,64]. At a structural level, more recent work has demonstrated that nucleation of PAA MTs depends on the prior formation of the cytokinetic actin ring (CAR) [67]. Interestingly, this work also demonstrated that the PAA in turn has a role in stabilizing the position of the CAR during extended delays in cytokinesis. The mechanism(s) by which this occurs are unknown, but they could involve minus end-directed MT motors transporting CAR-stabilizing components to the cell division site. More generally, these experiments highlight the fact that, as with astral MTs, we still don’t know the precise function of PAA MTs, as they are completely absent from _mto1_Δ mutants, which are viable (see below). PAA MTs have been hypothesized to play a role in keeping daughter nuclei away from each other and from the division site during mitosis, and also to contribute to setting up the initial state of MT organization for the next interphase [33].
A separate question relating to the eMTOC is how it is broken down at the end of mitosis, as septation proceeds, and how this relates to the appearance of iMTOCs in the next cell cycle. Although relatively little is known about eMTOC disassembly, it is likely to involve the action of molecular chaperones of the hsp70 family**,** because mutations in rsp1p, an hsp70-associated J-domain protein, lead to defects in eMTOC breakdown [105]. J-domain proteins act to promote the function of hsp70 chaperones in disassembling large protein complexes [15,95]; accordingly, in rsp1 mutants, large fragments of eMTOC material still capable of nucleating MTs leave the cell division site at the end of mitosis and diffuse in the cytoplasm in the subsequent cell cycle, with ensuing defects in interphase cytoplasmic MT organization.
Specialization in mating and meiosis
While a detailed discussion of cytoskeletal reorganization in meiosis is beyond the scope of this review, it is worth pointing out two distinct features of MTs in mating and/or meiotic cells. First, in mating cells, there is good evidence for a mating-specific MTOC at the tips of mating-projections [70]. The molecular basis for this remains largely unexplored. Later, after mating and cell-fusion have produced a zygote, the cytoplasmic MTs assume a different organization. Specifically, during meiotic prophase, MTs are nucleated exclusively from the SPB and are involved in so-called “horsetail” oscillatory movements of the prophase nucleus, which are thought to depend on cortical dynein pulling on the MTs [19,97]. Recent work has identified a meiosis-specific coiled-coil protein, mcp6p/hrs1p, which is localized to the SPB and is critical for the consolidation of MTOC activity to the SPB [74,84]. This will be an interesting area to follow.
The molecules of microtubule nucleation
The γ-TuC
While purified αβ-tubulin dimer can assemble into microtubules in vitro, microtubule nucleation in vivo nearly always involves the γ-tubulin complex (γ–TuC), a large (~2 MDa) specialized complex made up of several distinct proteins, including γ-tubulin [29,41,62,79]. The γ-TuC is best characterized biochemically in higher eukaryotes, where it is known as the γ-tubulin ring complex (γ-TuRC), because in the electron microscope it has a lock-washer structure with a diameter roughly that of a microtubule. Current models suggest that the γ-TuRC acts as a direct template for microtubule assembly, functioning to decrease the critical concentration for tubulin polymerization. The γ-TuRC is thought to contain probably six or seven copies of a subcomplex called the γ-tubulin small complex (γ-TuSC), which consists of two copies of γ-tubulin and one copy each of the γ-TuC proteins GCP2 and GCP3 [60,63]. Additional proteins in the complete γ-TuRC include the proteins GCP4, GCP5, GCP6 and GCP-WD [21,30,31,34,49,51,59,94,99]. Common sequence motifs are found in GCP2, GCP3, GCP4, GCP5 and GCP6, but their function is unknown [30,59].
Fission yeast homologs of γ-tubulin, GCP2 and GCP3 are known as gtb1p/tug1p, alp4p and alp6p, respectively [37,83,91]. All of these genes are essential for viability, almost certainly because of defects in mitotic spindle formation. Inferences relating to their role in cytoplasmic MT nucleation are largely derived from phenotypes of temperature-sensitive mutants. Such mutants typically show very long bundles of interphase MTs, which often curve around the cell tip [66,91]. In instances where MT nucleation has been studied more directly, for example, in alp4-1891 heat-sensitive mutants at a semi-permissive temperature, nucleation of both cytoplasmic astral MTs and PAA MTs appears to be compromised [103].
Subsequent work has identified fission yeast homologs of two γ-TuRC-specific proteins (i.e., those that are in the γ-TuRC but not the γ-TuSC) [25,93]. The homolog of GCP4 is gfh1p and the homolog of GCP6 is alp16p; similar homologs have not been found in budding yeast. The sequence similarity of gfh1p and alp16p to GCP4 and GCP6 (respectively) is remarkably low, in contrast to the high similarity between fission yeast and higher eukaryotic components of the γ-TuSC. Interestingly, neither _gfh1_+ nor _alp16_+ is an essential gene; this not only has implications for how they must be functioning in the γ-TuC but also makes it possible to analyze phenotypes of deletion mutants.
In both _gfh1_Δ and _alp16_Δ mutants, interphase microtubule organization is altered, with longer MTs bundles that often curl around cell tips, in many ways similar to temperature-sensitive mutants of gtb1+, _alp4_+ and alp6+. Less is known, however, as to whether these proteins have specific functions in MT nucleation. The initial characterization of _gfh1_Δ mutants [93] suggested that astral MTs fall off the SPB in mutants, implicating gfh1p in connecting the γ-TuC to the SPB. However, in the lab of one of us, recent work with these and other mutants has found no such defect (A. Anders and K.E.S., unpublished data). This same work also identified a novel fission yeast gene, mod21+, as a distant homolog of higher eukaryotic GCP5, and specifically assayed MTOC activity in _gfh1_Δ, _mod21_Δ and _alp16_Δ single mutants as well as triple deletion mutants. Overall, these mutants were found to have reduced MT nucleation specifically at iMTOCs, while nucleation of astral MTs and PAA MTs remained intact (A. Anders and K.E.S., unpublished data). Whether this reduction in iMTOC activity is the primary cause of the abnormal MT distributions observed in these mutants is not yet clear (see below).
Mto1p and mto2p
A major challenge in studying cytoplasmic MT organization in fission yeast is to understand how different types of cytoplasmic MTOCs can form and/or be active at different places and times during the cell cycle. In the last two years, two novel proteins, mto1p and mto2p, have been identified by several groups and shown to play a critical role in MT nucleation from cytoplasmic MTOCs [40,75,76,92,93,103]. Mto1 may have distantly-related homologs in higher eukaryotes [76]. Neither mto1p nor mto2p is required for mitotic spindle assembly, and neither is an essential gene, but deletion of either gene severely affects cytoplasmic MT organization. In _mto1_Δ mutants neither astral MTs nor PAA MTs are present, and interphase MTs are aberrantly bundled and often curve around cell tips (see below). In various assays for MT nucleation in vivo, _mto1_Δ mutants fail to nucleate any MTs in the cytoplasm; in fact, the only reason _mto1_Δ interphase cells have cytoplasmic MTs at all is that MTs nucleated inside the cell nucleus can “escape” into the cytoplasm [76,103]. In _mto2_Δ mutants, there is nearly no PAA, which makes the CAR unstable, but there are astral MTs [40,75,92]. This is consistent with observations that cytoplasmic MT nucleation can occur from the SPB in _mto2_Δ mutants in several different assays. Because both mto1p and mto2p localize to SPBs, to eMTOCs and to MT- and nuclear envelope-associated satellites that are thought to represent iMTOCs (see above), all evidence suggests that they act directly at cytoplasmic MTOCs to promote MT nucleation.
Biochemical work has shown that Mto1 and Mto2 physically interact and can co-immunoprecipitate the γ-TuC, although the efficiency of co-immunoprecipitation is not very high [40,75,76,92,93]. Interestingly, Mto1p cannot co-immunoprecipitate the γ-TuC in _mto2_Δ mutants. Moreover, in both _mto1_Δ and _mto2_Δ mutants, the failure to nucleate MTs from a given MTOC correlates with a failure to localize the γ-TuC complex to that MTOC [40,76,103]. This implies that mto1p and mto2p function by recruiting the γ-TuC to prospective MTOCs. Because the in vivo analyses show that _mto1_Δ phenotypes are more severe than _mto2_Δ phenotypes, it could be proposed that Mto1 may be the “major” protein interacting with the γ-TuC, and that Mto2 regulates the ability of Mto1 to interact with the γ-TuC [75]. However, such a model also requires that Mto1 at the SPB can function without Mto2. It is thought that a likely paralog of mto1p, pcp1p, may play a role analogous to mto1p in the assembly of mitoic spindle microtubules [23,76].
How to make an MTOC?
While a flurry of recent work has provided a solid framework for understanding how cytoplasmic MT nucleation is regulated in fission yeast, many new questions emerge. In particular, if spatially restricted MT nucleation in fission yeast depends on localization and/or activity of the γ-TuC, which in turn is recruited to MTOCs by mto1/2, it is now important to understand what controls the spatial and temporal localization of mto1p and mto2p. In addition, with regard to the γ-TuC itself, the function of gfh1p, mod21p and alp16p in regulating γ-TuC architecture and/or activity remains to be elucidated. Here, and also in relation to other questions, we need a better understanding of the molecular interactions occurring among the different components regulating MT nucleation. Particularly important are those interactions involving mto1p, mto2p and the γ-TuC. We don’t know whether mto1p and mto2p merely recruit the γ-TuC to prospective MTOCs, or if they also activate the γ-TuC, nor do we understand the specific role of mto2p in this process. More dedicated biochemical approaches will be important for addressing these questions.
B. Regulating Microtubule dynamics at the plus-end
At the other end of MTs, away from MTOCs and MT minus-ends, are MT plus-ends, which merit attention for two reasons. First, plus-ends play a role in signalling cell-polarity information to the cell cortex, mainly via the protein tea1p, which is deposited at the cortex at cell tips after being targetted there by an association with growing MT plus-ends [4,55,78,81,82]. The role of tea1p in cell polarity is outside the scope of this review and will not be discussed here [14,52,86]. Second, and common to all eukaryotic cells, is that MT plus-ends are the main sites where control of MT dynamics takes place, and proper regulation of dynamics is necessary for the generation of the stereotyped cytoplasmic MT arrays seen in fission yeast.
Dynamic instability and +TIPs
In fission yeast, as in most eukaryotic cells, MTs exhibit a behavior termed dynamic instability, with intervals of MT growth and MT shrinkage punctuated by “catastrophe” and “rescue” transitions [18]. In wild-type cells, the most commonly observed behavior is one in which MTs grow towards cell tips, pause briefly, and then initiate catastrophe and MT shrinking [20]. There is some in vivo evidence that in wild-type cells, dynamic instability transitions of individual MTs within bundles (e.g., from growth to shrinkage) are independent of other individual MTs [73], but for technical reasons this issue has not yet been investigated more exhaustively; further analysis in mutant strains would be particularly interesting. For the most part, recent analysis has focused on the dynamic behavior of MT bundles rather than that of individual MTs, as this is easier to analyse technically, although it may be a slight oversimplification.
In the last few years, many proteins have been identified that are associated with growing MT plus-ends, either directly or indirectly; these have been found in a broad range of eukaryotic cells and are often referred to as “+TIPs” (plus-end tracking proteins) [1,13]. The +TIP designation is conferred purely on the basis of association with MT ends in vivo rather than on functional or sequence-specific criteria, and thus +TIPs include several different protein families (for example, according to this definition, tea1p qualifies as a +TIP, although it is not obviously conserved across eukaryotes). In addition, the mechanisms by which +TIPs associate with plus-ends may be diverse, including MT motor-driven transport, co-assembly with tubulin dimer and/or binding to specific tubulin conformations at MT plus ends, and “hitch-hiking” by binding to other +TIPs [1,13].
In fission yeast, at least two +TIPs conserved in eukaryotic cells have been specifically implicated in regulating MT dynamics, most likely acting at a level very close to the MT polymer itself. One of these, tip1p, is the fission yeast homolog of mammalian CLIP-170 [9,69], and phenotypic analysis suggests that tip1p is important for suppressing MT catastrophe. In wild-type cells, contact between growing MT plus-ends and the cortex leads to catastrophe only when the contact occurs at cell tips, such that MT growth continues when a growing MT contacts the cortex in the middle regions of the cell. By contrast, in _tip1_Δ cells, MT-cortical contact leads to MT catastrophe regardless of where in the cell the contact is made [9]. As a result, _tip1_Δ mutants have shorter MT bundles under steady-state growth conditions, with deleterious consequences for microtubule-mediated regulation of cell polarity. A second protein, mal3p, is the fission yeast homolog of mammalian EB proteins [5,43,87]. Phenotypic analysis of _mal3_Δ mutants indicates that, like tip1p, mal3p plays a role in suppressing MT catastrophe. However, the MT-catastrophe phenotype of _mal3_Δ mutants is more severe than that of _tip1_Δ mutants; in _mal3_Δ mutants, MTs initiate catastrophe even before reaching the cell cortex [10]. Mal3p and tip1p physically interact, and mal3 is required for the proper +TIP localization of tip1p. An additional +TIP in the system, the kinesin-like protein tea2p, is required for transport of tip1p to MT plus ends [7,11]. As a result, _tea2_Δ cells also have shorter MTs at steady-state [8].
While tip1p and mal3p are likely among the most important +TIPs regulating MT dynamic instability, a great deal remains to be learned about how they work at a molecular level, and figuring this out will be a challenge. In particular, it is apparent from work in several different eukaryotic systems that many of the various +TIPs physically interact with each other, and with tubulin, in multiple ways [1]. This suggests that a complete understanding of +TIP function will not be represented as a simple linear pathway. Rather, a network model of functional interactions may be more appropriate, and this also appears to be the case in fission yeast [10,11,22]. In principle, relative to higher eukaryotes, fission yeast might represent a simplified system with which the complexities of +TIP mechanisms can be unravelled more easily. However, there are several striking differences in the behavior of at least some +TIPs in higher eukaryotes relative to the yeasts (both fission yeast and budding yeast, although we do not describe budding yeast here). For example, in higher eukaryotic cells, the association of proteins such as CLIP-170 with MTs assumes the form of a “comet tail”, often several microns long [69], whereas in fission yeast, the association of tip1p with MT plus ends is more punctate [9]. This may reflect not only differences in dissociation kinetics after targetting but also differences in how the proteins are initially targeted to plus-ends. In fission yeast, tip1p association with MT plus-ends is thought to involve primarily tea2p kinesin-mediated transport [11], whereas in higher eukaryotes, association of CLIP-170 with plus-ends may occur via coassembly with tubulin [1,3,24]. Another significant difference is that while both CLIP-170 and tip1p are involved in promoting MT growth overall, current evidence suggests that CLIP-170 acts primarily by promoting MT rescue [3,44], whereas tip1p acts by suppressing catastrophe [9].
Depending on one’s point of view, such differences might suggest that there is no single universal mechanism by which +TIPs regulate MT dynamics, and that apparent +TIP “homologs” might regulate MT dynamics in fundamentally different ways in different systems. An alternative view, however, would be that in the context of a network of multiple protein-protein interactions, at least some apparent differences in the mechanistic roles of homologous +TIPs are actually the consequences of quantitative changes in the binding constants of individual protein-protein interactions. That is, depending on which specific protein-protein interactions predominate in a given experimental system under physiological conditions, certain nodes or sub-pathways within a possible universe of pathways could be favored at the expense of others. To gain further understanding in this area will require much more detailed assessments of how specific protein-protein interactions and/or modifications of +TIPs contribute to their function, not only in vivo but also using in vitro reconstitution assays involving purified components [3,24]. Finally, a largely unexplored area in the field is the exact relationship between the cell-polarity signalling functions of MTs and the control of plus-end dynamics by +TIPs, as there is almost certainly molecular “cross-talk” between this two phenomena [7,22].
How do nucleation proteins contribute to MT dynamics?
While we know relatively little about the detailed mechanisms by which +TIPs such as tip1p and mal3p regulate MT plus-end dynamics, another area of MT dynamic instability regulation remains even more mysterious and more difficult to address experimentally. One of the most obvious features in many, and perhaps all, mutants affecting MT nucleation is that MT dynamics and behavior are altered. MTs often appear more strongly bundled in mutants, and bundles can persist in vivo for long periods of time, often curling around cell tips, and in some cases this also leads to oscillations of the SPB [25,40,54,66,75,76,85,91,92,93,103] (A. Anders and K.E.S., unpublished data). In addition, more careful analyses of MT behavior in several mutants have indicated unusual “treadmilling” of MT polymer within MT bundles, deviating from conventional dynamic instability behavior [40,103].
At present, nearly nothing is known about the mechanistic basis for these differences in MT dynamics, and here we would mainly want to point out that quite different views are possible. One end of the spectrum could be represented by a “purely phenotype-driven view”, that proteins conventionally thought to act exclusively in MT nucleation also play additional, direct roles in modulating bundling and/or plus end dynamics—that is, what is observed in vivo is a direct reflection of protein function. While there is no strong evidence for this view per se, it is noteworthy that MT-associated iMTOC satellites (see above) are motile within cells [40,75,76,103], and thus could be imagined to occasionally associate with MT plus ends and regulate dynamic instability transitions. Mechanistically, however, this may be hard to envision, because if anything, nucleation proteins might be expected specifically to associate with minus-ends rather than plus-ends, as in other systems, the γ-TuC can bind to and cap the MT minus-end [96].
At the opposite end of the spectrum is what could be termed a “systems-driven view”, that the effects on MT behavior observed in nucleation mutants are largely or completely indirect and due to a wide range of factors. For example, theoretical considerations suggest that under dynamic instability conditions, having a very small number of nucleation sites may lead to an increase in the partitioning of total cellular tubulin into tubulin-dimer relative to polymer phase [57]. Such an increase could in theory act to suppress catastrophe, although it might also be expected to increase growth rates. Another possibility is that factors regulating MT plus-end dynamics normally need to be loaded at MT minus-ends during nucleation [7] (with subsequent transport to plus-ends; see above), and this might not occur properly in nucleation mutants. Yet another mechanism closely linked to nucleation is the possibility that in wild-type cells, minus-ends not anchored at the SPB would normally be capped by nucleation complexes, and if this were not to occur, minus-ends could be free to further elongate and fall under the malign influence of MT bundling factors (see below), becoming less likely to undergo catastrophe as a result. Following on from the idea that MT bundling might be an indirect route to modulating dynamics, one can imagine more generally that if there were only one nucleation site in the cell (for example, the SPB), then all nucleated MTs would be near neighbors, and more likely to become overbundled and artificially stabilized.
We tend to favor the view that indirect effects may be behind the differences in MT dynamics seen in nucleation mutants, if only because there are relatively little data to support a direct role for nucleation proteins in regulating dynamics, while there are clearly many possible ways to achieve indirect effects. However, to date, both views are ultimately derived only from phenotypic observations in vivo, and until such work is complemented by more mechanistic studies, it is important to keep an open mind. As with studies on MT nucleation and the regulation of MT plus-end dynamics by +TIPs, in vitro systems reconstituting MT dynamics may help significantly to illuminate these questions.
Organizing Microtubule Bundles
Here we focus on a third area of cytoplasmic MT organization, the nature of interphase MT bundles and their polarity. Interphase fission yeast typically have three to four bundles of MTs running along the long axis of the cell [20,89]. The (average) number of microtubules per bundle is still uncertain, as definitive determinations require pain-staking electron-microscopy reconstructions; however, one study of bundle cross-sections suggests 2-5 MTs per bundle [12]. This would be roughly consistent with observations of GFP-tubulin fluorescence intensity from many workers, although where microtubules begin and end within any given bundle is still an open question, especially as we know that iMTOC satellites can be present throughout the length of MTs (see section A, above). The MTs in each bundle are generally organized such that the growing plus ends are pointed toward the opposite cell tips, and the stable minus ends are bundled together at the cell center [12,20,89], effectively leading to an antiparallel and symmetrical linear MT array. In fission yeast, this organization is important for two reasons. First, the MT arrays generate opposing pushing forces to position the interphase nucleus at the cell center, the future site for the placement of the cell division plane [17,89]. Second, as described above, the MT plus ends, through regulated delivery of polarity determinants such as tea1p [7,10,11], define the linear growth axis of the cell [55,78]. More generally, the organization of interphase MTs into antiparallel linear arrays in fission yeast may give insights into how higher-ordered cytoskeletal structures can be organized in diverse cell types for different functions. Recent work highlights two key molecules involved in the organization of fission yeast interphase MT arrays: the MT-associated protein (MAP) ase1p and the kinesin-14 motor klp2p.
Bundlers and sliders
Ase1p, which is conserved in fungi, animals and plants [50,80,102], localizes to the overlapping minus ends of fission yeast interphase MT bundles [48,98]. In _ase1_Δ cells, the interphase MTs are disorganized and showed no bundled minus ends at the cell center [48]. The kinesin-14 motor klp2p belongs to the class of minus end-directed MT motors (see [47] for kinesin nomenclature) and localizes as a relatively small number of motile dots (probably 10-15 per cell) along the interphase MT arrays [90]. In _klp2_Δ cells, no sliding of newly created MTs on preexisting MTs can occur, leading to unfocused regions of MT overlap at the cell center and deviations from the normal distribution of MT plus-ends towards cell tips and MT minus-ends towards the cell middle, as judged by the transport of the +TIP tea1p [12]. These results suggest that ase1p serves as a MT bundler and klp2p serves as a MT slider, and that the interplay between bundlers and sliders dictates the organization of MTs into antiparallel linear arrays. Based on these results and more recent unpublished data, a general hypothetical model for how fission yeast builds an interphase MT bundle can be proposed (Fig. 2). We discuss implications of this model and some possible future directions of research.
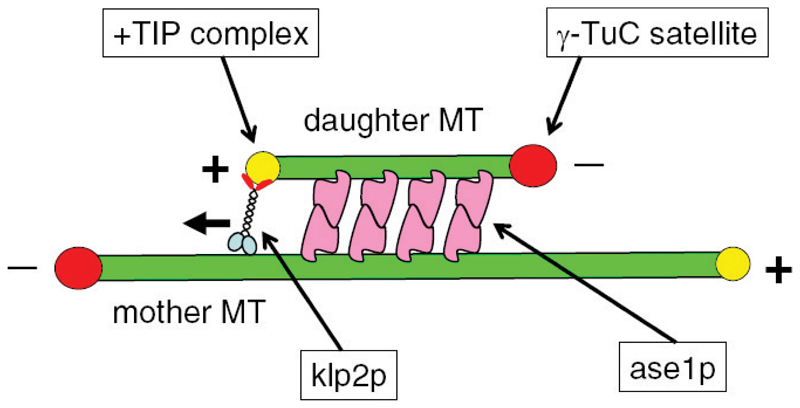
Figure 2. Model of interphase microtubule bundling in fission yeast.
A representation of how microtubules (MTs), motors, and microtubule-associated proteins (MAPs) may be necessary and sufficient to bundle and slide two MTs together into an antiparallel MT array. The model includes the following steps: 1) A cytoplasmic γ-tubulin complex (γ-TuC) satellite is recruited to the lattice of a preexisting mother MT. The mechanism of recruitment is not known. 2) The γ-TuC satellite nucleates a new daughter MT. The mechanism for recruitment-dependent nucleation is not known. 3) Ase1p bundles and stabilizes the antiparallel arrangement of daughter-mother MTs. Ase1p binding would be dynamic, so the bundling and stabilizing activities would still allow for MT sliding. 4) Klp2p is recruited to the growing plus end tip of the daughter MT, where it “pulls” the daughter MT toward the minus end of the mother MT. Pulling effectively slides the daughter and mother MT relative to each other. Sliding is attenuated as daughter MT continues to grow and new ase1p is recruited to the growing overlap region between daughter-mother MTs. The mechanism of klp2p attachment to the MT plus end tip is not known. 5) When klp2p reaches the end of the mother MT, no further sliding occurs, and the length of the daughter-mother overlap region is defined. 6) The daughter MT may then continue to grow beyond the mother’s minus-end, establishing an antiparallel and symmetric MT bundle with minus-ends bundled together at the middle, and plus-ends extending toward the cell tips (see Fig. 1).
According to the model, once γ-TuC satellite iMTOCs have nucleated a new daughter MT on a preexisting mother MT [40], mother and daughter MTs would be further organized into functional patterns primarily by the concerted action of ase1p and klp2p. Emerging data suggests that ase1p could function as a homo-dimer to preferentially bundle mother and daughter MTs into an antiparallel linear array—that is, with the mother MT having its minus end at the nuclear envelope and its plus end facing one of the cell tips, and the daughter MT having its minus end facing one of the cell tips and its plus end pointing toward the mother MT minus end (M. Janson and P.T.T., unpublished data). Klp2p would be localized at the plus end tip of the growing daughter MT (i.e., acting like a +TIP) and move along the mother MT toward its (i.e., the mother’s) minus end, thus “pulling ” the trailing daughter MT toward the minus end of the mother MT. Once the klp2p-coupled plus end of the daughter MT reaches the end of the (stable) minus end of the mother MT, no further sliding between daughter and mother MTs would occur, and a stable overlapping region between mother and daughter MTs would now be defined and fixed at the nuclear region, even if the plus-end of the daughter MT were to extend beyond the minus-end of the mother MT. Accordingly, in _ase1_Δ cells, no stable overlapping regions between daughter and mother MTs would be established, while in _klp2_Δ cells, no transport of the daughter MT would occur, in which case ase1p might continue to bundle daughter and mother MTs into potentially larger antiparallel overlapping regions.
This speculative model raises some interesting points for future experiments and also provokes more questions. The first point is that a three-component system involving MT-nucleation, the bundler ase1p and the slider klp2p may be necessary and sufficient to organize antiparallel linear arrays of MTs. Second, the length of the overlapping daughter-mother MT region would be determined when klp2p reaches the end of the minus end of the mother MT. Third, the localization of ase1p to regions of MT overlap would not be dependent on MT motors. We discuss these issues below.
Simulating the antiparallel-MT generator
A three-component system is sufficiently simple to be amenable to computer modeling and simulation. Past attempts on modeling MT arrays have mostly focused on the mitotic spindle, specifically the midzone, where mitotic kinesins are localized to organize the bipolar symmetric spindle and to slide the spindle apart (see [58] for review). These models did not include possible contribution of MAPs such as ase1p. Some recent attempts at simulation suggested that MT-nucleation, bundling, and sliding are necessary and sufficient for organizing antiparallel arrays of MTs [61]. Further advances on simulation will have to include the contributions of both motors and MAPs, and be able to help predict, for example, the average lengths of the overlapped MT regions.
Regulating the overlap zone
Having klp2p at the plus-end of the daughter MT would ensure that when the daughter plus-end reaches the minus end of the mother MT, there would necessarily be a remaining overlap region between the mother and daughter MTs (Fig. 2). However, several questions arise in relation to the length and lifetime of overlap zones. For example, as the daughter MT elongates by new tubulin subunit addition (i.e., before reaching the mother’s minus-end), progressively more ase1p would be expected to be recruited to the mother-daughter MT overlap region. Such an increase in ase1p-dependent bundling could potentially counteract the klp2p-driven movement of a daughter MT, slowing it down (see, for example, Fig. 8B of [40]. Thus the length of the overlap region might be regulated by a complicated interplay between tubulin polymerization kinetics, ase1p binding, and the performance of the klp2p MT motor under increased load. Another outstanding question is how klp2p would remain attached to the plus-end of a growing daughter MT as it elongates, while simultaneously moving the daughter MT as cargo. Possible candidates that may play a role in attaching klp2p to the plus end tip of the daughter MT include +TIP proteins such as mal3p, tip1p and/or tea1p. Further investigations into the physical interactions among +TIPs and how they associate with MT lattice conformations may yield more insights into how klp2p could simultaneously bind and pull on a dynamic MT. Yet another major challenge would be to understand what regulates the number and spacing of daughter MTs on the mother MTs (partly a nucleation question), as well as how turnover of daughter and mother MTs is regulated, especially in the overlap zone. Although the general picture of MT dynamic instability involves bi-directional switching between persistent states of growth and shrinkage, it is not really clear whether conventional “rescue” actually occurs in fission yeast, as some experiments suggest that once a switch from growth to shrinkage occurs, MTs shrink completely [9,10,20,89]. While technical limitations to imaging make this difficult to judge with complete certainty, such results suggest that there would be a constant need for recruitment of daughter MTs to the center of the bundle to replace the disassembling mother MTs [40,100]. Thus, the lifetime of a MT bundle could be limited by the rate of recruitment of new daughter MTs to the overlap zone, as part of an iterative process in which older daughters themselves later become new mothers, etc.
Bundling independent of motors?
It has been proposed for mammalian cells that the kinesin-4 motor KIF4 may carry PRC1, the mammalian ase1p homolog, to sites of MT overlap at the spindle midzone [46,101]. Emerging work in fission yeast suggest an alternative mechanism to the notion that motors carry MAPs to region of interest. Specifically, there is evidence that ase1p can by itself find regions of MT overlap, because in the absence of klp2p, ase1p still localizes specifically to the mother-daugher MT overlapping region (M. Janson and P.T.T., unpublished data). From a structural perspective, this could imply that ase1p homo-dimerization creates a relatively rigid molecule, promoting mostly antiparallel bundling, and also possibly that binding of ase1p dimer to MTs may be cooperative, controlled by allostery. As a MT bundler, perhaps ase1p has the ability to quickly scan the MT lattice until it meet two MTs in close proximity, allowing it specifically to organize regions of antiparallel MT overlap. Such a scanning or “skating” mechanism has been recently proposed for the non-walking kinesin MCAK and for dynein/dynactin [16,36].
Conclusions
It is only in the last few years, since the adoption of new methods such as live-cell imaging of GFP-tubulin, that it has been possible to move towards a more complete understanding of cytoplasmic MT organization in fission yeast. From this brief survey of proteins involved in the three processes of MT nucleation, plus-end dynamics, and bundling, it is clear that the systems regulating MT behavior are complex and also closely interconnected. At the same time, recent progress suggests that the interesting problems are experimentally tractable. In particular, we believe that several of the key molecules in each of the three processes discussed have now been identified. In the case of cytoplasmic MT nucleation, mto1p and mto2p act to direct the γ-TuC to prospective MT sites. In the regulation of MT plus-end dynamics, mal3 and tip1p likely play very prominent roles. In the formation of antiparallel MT bundles, ase1p and klp2p appear to be the major players. Further investigations of how these proteins work in a physiological context should continue to yield new insights.
Acknowledgements
We thank Andreas Anders, Marcel E. Janson, Isabelle Loiodice and Itaru Samejima for insightful discussions. Work in the laboratory of P.T.T is supported by the National Institutes of Health (RO1-GM70899). Work in the laboratory of K.E.S. is supported by the Wellcome Trust. K.E.S. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences.
References
- 1.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th edition. Garland Science; 2002. [Google Scholar]
- 3.Arnal I, Heichette C, Diamantopoulos GS, Chretien D. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr Biol. 2004;14:2086–95. doi: 10.1016/j.cub.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 4.Behrens R, Nurse P. Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J Cell Biol. 2002;157:783–93. doi: 10.1083/jcb.200112027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–28. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergen LG, Borisy GG. Head-to-tail polymerization of microtubules in vitro. Electron microscope analysis of seeded assembly. J Cell Biol. 1980;84:141–50. doi: 10.1083/jcb.84.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning H, Hackney DD, Nurse P. Targeted movement of cell end factors in fission yeast. Nat Cell Biol. 2003;5:812–8. doi: 10.1038/ncb1034. [DOI] [PubMed] [Google Scholar]
- 8.Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol. 2000;151:15–28. doi: 10.1083/jcb.151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 10.Busch KE, Brunner D. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr Biol. 2004;14:548–59. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Busch KE, Hayles J, Nurse P, Brunner D. Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev Cell. 2004;6:831–43. doi: 10.1016/j.devcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Carazo-Salas RE, Antony C, Nurse P. The kinesin Klp2 mediates polarization of interphase microtubules in fission yeast. Science. 2005;309:297–300. doi: 10.1126/science.1113465. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho P, Tirnauer JS, Pellman D. Surfing on microtubule ends. Trends Cell Biol. 2003;13:229–37. doi: 10.1016/s0962-8924(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 14.Chang F, Peter M. Yeasts make their mark. Nat Cell Biol. 2003;5:294–299. doi: 10.1038/ncb0403-294. [DOI] [PubMed] [Google Scholar]
- 15.Craig EA, Huang P, Aron R, Andrew A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 16.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8:264–70. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 17.Daga RR, Chang F. Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci U S A. 2005;102:8228–32. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 19.Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–12. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- 20.Drummond DR, Cross RA. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr Biol. 2000;10:766–75. doi: 10.1016/s0960-9822(00)00570-4. [DOI] [PubMed] [Google Scholar]
- 21.Fava F, Raynaud-Messina B, Leung-Tack J, Mazzolini L, Li M, Guillemot JC, Cachot D, Tollon Y, Ferrara P, Wright M. Human 76p: A new member of the gamma-tubulin-associated protein family. J Cell Biol. 1999;147:857–68. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feierbach B, Verde F, Chang F. Regulation of a formin complex by the microtubule plus end protein tea1p. J Cell Biol. 2004;165:697–707. doi: 10.1083/jcb.200403090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flory MR, Morphew M, Joseph JD, Means AR, Davis TN. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 2002;13:47–58. [PubMed] [Google Scholar]
- 24.Folker ES, Baker BM, Goodson HV. Interactions between CLIP-170, tubulin, and microtubules: implications for the mechanism of Clip-170 plus-end tracking behavior. Mol Biol Cell. 2005;16:5373–84. doi: 10.1091/mbc.E04-12-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita A, Vardy L, Garcia MA, Toda T. A Fourth Component of the Fission Yeast gamma-Tubulin Complex, Alp16, Is Required for Cytoplasmic Microtubule Integrity and Becomes Indispensable When gamma-Tubulin Function Is Compromised. Mol Biol Cell. 2002;13:2360–73. doi: 10.1091/mbc.02-01-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gachet Y, Reyes C, Goldstone S, Tournier S. The fission yeast spindle orientation checkpoint: a model that generates tension? Yeast. 2006 doi: 10.1002/yea.1410. XX: XXX-XXX. [DOI] [PubMed] [Google Scholar]
- 27.Gachet Y, Tournier S, Millar JB, Hyams JS. A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature. 2001;412:352–5. doi: 10.1038/35085604. [DOI] [PubMed] [Google Scholar]
- 28.Gachet Y, Tournier S, Millar JB, Hyams JS. Mechanism controlling perpendicular alignment of the spindle to the axis of cell division in fission yeast. EMBO J. 2004;23:1289–1300. doi: 10.1038/sj.emboj.7600156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunawardane RN, Lizarraga SB, Wiese C, Wilde A, Zheng Y. gamma-Tubulin complexes and their role in microtubule nucleation. Curr Top Dev Biol. 2000;49:55–73. doi: 10.1016/s0070-2153(99)49004-0. [DOI] [PubMed] [Google Scholar]
- 30.Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, Zheng Y. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J Cell Biol. 2000;151:1513–24. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunawardane RN, Martin OC, Zheng Y. Characterization of a new gammaTuRC subunit with WD repeats. Mol Biol Cell. 2003;14:1017–26. doi: 10.1091/mbc.E02-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111:1603–12. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- 33.Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–57. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- 34.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol. 2006;172:505–15. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heitz MJ, Petersen J, Valovin S, Hagan IM. MTOC formation during mitotic exit in fission yeast. J Cell Sci. 2001;114:4521–32. doi: 10.1242/jcs.114.24.4521. [DOI] [PubMed] [Google Scholar]
- 36.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–9. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 37.Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- 38.Huffaker TC, Thomas JH, Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs CW, Adams AE, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–26. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Job D, Valiron O, Oakley B. Microtubule nucleation. Curr Opin Cell Biol. 2003;15:111–7. doi: 10.1016/s0955-0674(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 42.Keating TJ, Borisy GG. Centrosomal and non-centrosomal microtubules. Biol Cell. 1999;91:321–9. [PubMed] [Google Scholar]
- 43.Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell. 2005;16:5334–45. doi: 10.1091/mbc.E05-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG. Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol. 2002;159:589–99. doi: 10.1083/jcb.200208058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004;14:R722–30. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 46.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–48. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–68. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–47. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 50.Mao G, Chan J, Calder G, Doonan JH, Lloyd CW. Modulated targeting of GFP-AtMAP65-1 to central spindle microtubules during division. Plant J. 2005;43:469–78. doi: 10.1111/j.1365-313X.2005.02464.x. [DOI] [PubMed] [Google Scholar]
- 51.Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gammaTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–87. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–91. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Masuda H, Hirano T, Yanagida M, Cande WZ. In vitro reactivation of spindle elongation in fission yeast nuc2 mutant cells. J Cell Biol. 1990;110:417–25. doi: 10.1083/jcb.110.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuda H, Toda T, Miyamoto R, Haraguchi T, Hiraoka Y. Modulation of Alp4 function in Schizosaccharomyces pombe induces novel phenotypes that imply distinct functions for nuclear and cytoplasmic gamma-tubulin complexes. Genes Cells. 2006;11:319–36. doi: 10.1111/j.1365-2443.2006.00946.x. [DOI] [PubMed] [Google Scholar]
- 55.Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–49. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 56.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 57.Mitchison TJ, Kirschner MW. Some thoughts on the partitioning of tubulin between monomer and polymer under conditions of dynamic instability. Cell Biophys. 1987;11:35–55. doi: 10.1007/BF02797111. [DOI] [PubMed] [Google Scholar]
- 58.Mogilner A, Wollman R, Civelekoglu-Scholey G, Scholey J. Modeling mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 59.Murphy SM, Preble AM, Patel UK, O'Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell. 2001;12:3340–52. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–74. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nedelec F. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J Cell Biol. 2002;158:1005–15. doi: 10.1083/jcb.200202051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oakley BR. gamma-Tubulin. Curr Top Dev Biol. 2000;49:27–54. doi: 10.1016/s0070-2153(99)49003-9. [DOI] [PubMed] [Google Scholar]
- 63.Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–33. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–73. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 65.Oliferenko S, Balasubramanian MK. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat Cell Biol. 2002;4:816–20. doi: 10.1038/ncb861. [DOI] [PubMed] [Google Scholar]
- 66.Paluh JL, Nogales E, Oakley BR, McDonald K, Pidoux AL, Cande WZ. A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol Biol Cell. 2000;11:1225–39. doi: 10.1091/mbc.11.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pardo M, Nurse P. Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science. 2003;300:1569–74. doi: 10.1126/science.1084671. [DOI] [PubMed] [Google Scholar]
- 68.Pardo M, Nurse P. The nuclear rim protein Amo1 is required for proper microtubule cytoskeleton organisation in fission yeast. J Cell Sci. 2005;118:1705–14. doi: 10.1242/jcs.02305. [DOI] [PubMed] [Google Scholar]
- 69.Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–27. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- 70.Petersen J, Heitz MJ, Hagan IM. Conjugation in S. pombe: identification of a microtubule-organising centre, a requirement for microtubules and a role for Mad2. Curr Biol. 1998;8:963–6. doi: 10.1016/s0960-9822(98)70397-5. [DOI] [PubMed] [Google Scholar]
- 71.Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell. 1998;9:1757–71. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rajagopalan S, Bimbo A, Balasubramanian MK, Oliferenko S. A potential tension-sensing mechanism that ensures timely anaphase onset upon metaphase spindle orientation. Curr Biol. 2004;14:69–74. doi: 10.1016/j.cub.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 73.Sagolla MJ, Uzawa S, Cande WZ. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J Cell Sci. 2003;116:4891–903. doi: 10.1242/jcs.00796. [DOI] [PubMed] [Google Scholar]
- 74.Saito TT, Tougan T, Okuzaki D, Kasama T, Nojima H. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J Cell Sci. 2005;118:447–59. doi: 10.1242/jcs.01629. [DOI] [PubMed] [Google Scholar]
- 75.Samejima I, Lourenco PC, Snaith HA, Sawin KE. Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol Biol Cell. 2005;16:3040–51. doi: 10.1091/mbc.E04-11-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawin KE, Lourenco PC, Snaith HA. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol. 2004;14:763–75. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 77.Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J Cell Biol. 1998;142:457–71. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sawin KE, Snaith HA. Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J Cell Sci. 2004;117:689–700. doi: 10.1242/jcs.00925. [DOI] [PubMed] [Google Scholar]
- 79.Schiebel E. gamma-tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr Opin Cell Biol. 2000;12:113–8. doi: 10.1016/s0955-0674(99)00064-2. [DOI] [PubMed] [Google Scholar]
- 80.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–28. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snaith HA, Samejima I, Sawin KE. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 2005;24:3690–9. doi: 10.1038/sj.emboj.7600838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snaith HA, Sawin KE. Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature. 2003;423:647–651. doi: 10.1038/nature01672. [DOI] [PubMed] [Google Scholar]
- 83.Stearns T, Evans L, Kirschner M. Gamma-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–36. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- 84.Tanaka K, Kohda T, Yamashita A, Nonaka N, Yamamoto M. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr Biol. 2005;15:1479–86. doi: 10.1016/j.cub.2005.07.058. [DOI] [PubMed] [Google Scholar]
- 85.Tange Y, Fujita A, Toda T, Niwa O. Functional dissection of the gamma-tubulin complex by suppressor analysis of gtb1 and alp4 mutations in Schizosaccharomyces pombe. Genetics. 2004;167:1095–107. doi: 10.1534/genetics.104.027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr Biol. 2005;15:1006–15. doi: 10.1016/j.cub.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 87.Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–6. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–6. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 89.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR. pkl1(+)and klp2(+): Two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell. 2001;12:3476–88. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vardy L, Toda T. The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 2000;19:6098–111. doi: 10.1093/emboj/19.22.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Venkatram S, Jennings JL, Link A, Gould KL. Mto2p, a novel fission yeast protein required for cytoplasmic microtubule organization and anchoring of the cytokinetic actin ring. Mol Biol Cell. 2005;16:3052–63. doi: 10.1091/mbc.E04-12-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Venkatram S, Tasto JJ, Feoktistova A, Jennings JL, Link AJ, Gould KL. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol Biol Cell. 2004;15:2287–301. doi: 10.1091/mbc.E03-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verollet C, Colombie N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol. 2006;172:517–28. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–71. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–64. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 97.Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–46. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–95. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang L, Keating TJ, Wilde A, Borisy GG, Zheng Y. The role of Xgrip210 in gamma-tubulin ring complex assembly and centrosome recruitment. J Cell Biol. 2000;151:1525–36. doi: 10.1083/jcb.151.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng L, Schwartz C, Wee L, Oliferenko S. The fission yeast transforming acidic coiled coil-related protein Mia1p/Alp7p is required for formation and maintenance of persistent microtubule-organizing centers at the nuclear envelope. Mol Biol Cell. 2006;17:2212–22. doi: 10.1091/mbc.E05-08-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci U S A. 2005;102:343–8. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci U S A. 2006;103:6196–201. doi: 10.1073/pnas.0506926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zimmerman S, Chang F. Effects of {gamma}-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol Biol Cell. 2005;16:2719–33. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zimmerman S, Daga RR, Chang F. Intra-nuclear microtubules and a mitotic spindle orientation checkpoint. Nat Cell Biol. 2004;6:1245–6. doi: 10.1038/ncb1200. [DOI] [PubMed] [Google Scholar]
- 105.Zimmerman S, Tran PT, Daga RR, Niwa O, Chang F. Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev Cell. 2004;6:497–509. doi: 10.1016/s1534-5807(04)00096-6. [DOI] [PubMed] [Google Scholar]