Microarray Analysis of Developing Arabidopsis Seeds (original) (raw)
Abstract
To provide a broad analysis of gene expression in developing Arabidopsis seeds, microarrays have been produced that display approximately 2,600 seed-expressed genes. DNA for genes spotted on the arrays were selected from >10,000 clones partially sequenced from a cDNA library of developing seeds. Based on a series of controls, sensitivity of the arrays was estimated at one to two copies of mRNA per cell and cross hybridization was estimated to occur if closely related genes have >70% to 80% sequence identity. These arrays have been hybridized in a series of experiments with probes derived from seeds, leaves, and roots of Arabidopsis. Analysis of expression ratios between the different tissues has allowed the tissue-specific expression patterns of many hundreds of genes to be described for the first time. Approximately 25% of the 2,600 genes were expressed at ratios ≥ 2-fold higher in seeds than leaves or roots and 10% at ratios ≥ 10. Included in this list are a large number of proteins of unknown function, and potential regulatory factors such as protein kinases, phosphatases, and transcription factors. The Arabidopsis arrays were also found to be useful for transcriptional profiling of mRNA isolated from developing oilseed rape (Brassica napus) seeds and expression patterns correlated well between the two species.
The major economic and food value of most agricultural products resides in their seeds and centuries of agricultural research have been directed at improving the qualitative and quantitative traits associated with seed products. Different major crop species produce seeds with very different compositions, which in large part reflect the proportions of the major storage components accumulated in the seeds. For example, Graminaceous species such as wheat, rice, and maize produce seeds that contain starch as the dominant component, whereas other crops produce seeds high in oil (e.g. rapeseed) or protein (e.g. soybean). Although the biosynthetic pathways responsible for accumulation of the major seed storage components are now largely defined (Ohlrogge and Jaworski, 1997; Eastmond and Rawsthorne, 2000), much less is understood about the mechanisms that determine the very different partitioning of seed reserves into the major storage components (Thomas, 1993).
The emergence of Arabidopsis as a major model system for plant science together with the development of extensive tools for its genetic and molecular dissection has led to major advances in understanding of many aspects of plant biology. Although a number of mutants in seed development (Franzmann et al., 1995) and in seed lipid biosynthesis (for review, see Ohlrogge et al., 1991; Katavic et al., 1995; Focks and Benning, 1998) are known, these represent only a few percent of the currently well-characterized Arabidopsis mutations. In a large part because of its very small seeds and the resulting technical difficulties, less effort has been focused on analysis of seed biology of Arabidopsis than for many other species. However, Arabidopsis as a Brassicaceae is an excellent model for major world oilseed crops such as oilseed rape (Brassica napus) to which it is closely related. Furthermore, because of facile and rapid methods to produce and analyze mutant and transgenic Arabidopsis, and the availability of a complete genome sequence, more extensive and rapid analysis of many aspects of seed biology can be conducted in Arabidopsis than in other species. As a component of such studies, and to take advantage of available Arabidopsis genetic and molecular tools, we have constructed microarrays based on 10,500 expressed sequence tags (ESTs) recently sequenced from an Arabidopsis developing seed cDNA library (White et al., 2000). These microarrays provide a tool to broadly analyze the expression of several thousand genes during seed development, to identify tissue-specific expression patterns, and to identify candidate genes for further more detailed analysis.
RESULTS
A Microarray from Developing Arabidopsis Seeds
From a cDNA library of developing Arabidopsis seeds, 27,568 clones were arrayed on filters and hybridized with probes specific for highly abundant transcripts (such as storage proteins) in Arabidopsis seeds. Over 10,000 clones, which showed no signal in this subtractive screening, were partially sequenced from their 5′ ends. Subsequent BLASTX and contig analysis condensed the number of these ESTs down to about 5,800 putative unique sequences. An approximate 30% of these sequences were not represented in the public Arabidopsis EST database (dbEST) as of October 1999, and >45% of these sequences had no significant similarity (BLAST score <100) to the entries in the GenBank protein database. This large number of potentially new sequences in part reflects the lack of study of Arabidopsis seeds by EST approaches and emphasizes the value of this cDNA set as an interesting resource for the discovery of novel gene functions. A more complete description of the generation of the seed-specific cDNA library, the sequencing project, and its analysis is given in White et al. (2000).
For microarray fabrication, a subset of 2,715 clones was selected from the 5,800 putative unique sequences. Some of these ESTs were very similar and are likely to represent the same gene. The number of unique genes represented on the arrays is therefore slightly less than 2,715. To monitor the expression pattern from as many genes involved in glycerolipid and carbohydrate metabolism as possible, 82 additional cDNA clones were collected that complemented the seed microarrays with most of the missing sequences from these pathways. In addition, a collection of 60 control DNAs was generated. The inserts of the three clone collections were amplified by PCR with vector-specific primers. PCR samples that yielded less than 0.2 mg/mL DNA or showed several DNA fragments were re-amplified or replaced with alternative clones. The PCR products were arrayed on and bound to poly-Lys-coated microscope slides. To increase the reliability of the detected signals, each PCR sample was spotted twice in two subarrays resulting in a total array of 7,680 data points. The identity of 37 randomly chosen DNA samples was confirmed by re-sequencing their PCR products used for microarray printing and comparing the obtained sequence results with the corresponding EST sequences in our database. In all 37 cases the sequences of the PCR samples matched with their original EST sequence. This sequence confirmation increases the confidence in the identity of the DNA elements on our microarrays and makes it unlikely that major errors in the selection of clones or sample plates occurred during sample preparation. Additional details of the microarray results from this study are available on-line at:http://www.bpp.msu.edu/Seed/SeedArray.htm.
Quality Control
To evaluate the reliability of the hybridization experiments, the microarrays contained several control elements. To detect the sensitivity limit and to have an additional control for balancing the intensities of the two channels, nine non-related human cDNA fragments were arrayed on the slides. The corresponding in vitro transcribed poly(A)+ RNA species were added to 1.0 μg of the plant tissue mRNA samples as internal standards in decreasing concentrations from 1.0 (1:1.0 × 10−3) to 0.01 ng (1:1.0 × 10−5; Fig.1). The lowest control RNA levels of 7.5 × 10−4 and 1.0 × 10−5 gave in most experiments fluorescence signal intensities higher than two times the local background. Similar detection limits of 1.0 × 10−5 (Ruan et al., 1998) and 5.0 × 10−5 (Schena et al., 1996) were detected by other groups. According to mRNA quantifications from Okamuro and Goldberg (1989) this detection limit corresponds to approximately one to two mRNA copies per cell.
Figure 1.
Microarray segments from repeated experiments. The two images show the same segments from two microarray hybridizations in false color presentation. In both experiments the arrays were cohybridized with fluorescence probes from seeds and leaves. A represents an experiment in which the leaf sample was labeled with Cy3 and the seed sample with Cy5. B shows the replicate experiment in which the fluorescence labels were incorporated in opposite orientation (leaf with Cy5; seed with Cy3). 12S and MDH indicate elements that contain DNA fragments coding for 12S seed storage protein (clone M10C10S) and plastidial malate dehydrogenase (clone M44F11), respectively. The white boxes highlight control elements localized within the given image segment. In the following they are explained always from the top to the bottom of the boxes. Specificity (cross hybridization) controls, Box 1 shows specificity controls consisting of three 365-bp FAD2 fragments with identical GC content; original FAD2 sequence, two synthetic forms with 90% and 80% identity; box 2 contains three different sequences coding for ferredoxins; as shown in Figure 1 and as described in “Materials and Methods,” additional controls monitored for nonspecific hybriclization carry over during and for mRNA integrity/probe length, from soybean with 63% identity to corresponding Arabidopsis sequence, from Impatiens with 66% identity, and from Arabidopsis. Amount of rRNA in mRNA and probe samples, Box 3 contains sequences coding for 25S rRNA and 18S rRNA from Arabidopsis. Carry-over of DNA during printing process, Box 4 includes a highly expressed sequence for Rubisco SSU and a negative control containing only 3× SSC, which were arrayed in this order from the same printing pin. Unspecific cross hybridization, Box 5 contains six PCR products from unrelated human cDNA sequences with the IDs 136643, 204716, 60027, 756944, 29328, and IB187. RNA and cDNA probe quality control, Box 6 contains 365-bp fragments from the 5′-, central, and 3′-regions of the FAD2 sequence and its full-length form of about 1,100 bp. Sensitivity control, Box 7 shows four elements containing different human sequences (IDs 1593605, 1020153, 1592600, and 1576490) for which in vitro transcribed poly(A)+ RNA was spiked into the RNA samples in concentrations of 1:10,000, 1:25,000, 1:50,000, and 1:75,000.
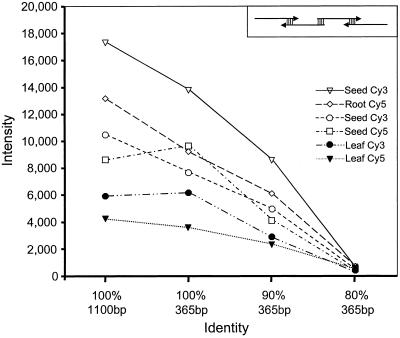
Many Arabidopsis genes belong to gene families, and therefore cross hybridizations between different members of gene families are an issue in cDNA based microarray experiments. Estimates of the extent of gene families in Arabidopsis range from 15% to 50% and over one-half of 64 proteins surveyed for lipid metabolism were found to be members of gene families (Mehkedov et al., 2000). To estimate the extent of possible cross hybridizations between related genes, the threshold of cross hybridization was detected in each experiment with several specificity controls. These controls included synthetic gene fragments and heterologous sequences from other plant species, which have decreasing sequence identities of 100% to 60% to three moderately expressed Arabidopsis genes. First, we synthesized and arrayed 365-bp synthetic fragments of the Arabidopsis FAD2 gene in three different forms of identical length and constant GC content of 48%, but decreasing nucleotide identities of 100%, 90%, and 80%. As shown in Figures 1 and 2, the 100% fragment gave comparably strong signals (generally within 80%–90%) to a 1.1-kbp PCR fragment from_FAD2_, indicating that a target length of 365 bp is sufficient for efficient probe binding in this technique. The 90% identity fragment gave approximately 50% weaker signals compared with the 100% form, whereas the 80% form showed almost no detectable signals suggesting a cross hybridization threshold under the conditions of these experiments between 80% to 90% identity. Cross reactions with other Arabidopsis transcripts are unlikely because for Arabidopsis, no genes are known that are closely related (>60%) to_FAD2_ (Okuley et al., 1994). The synthetic gene fragments were designed with evenly spaced mismatches. Two other specificity control sets consisted of four ferredoxin sequences and three acyl-ACP-desaturase sequences from other organisms. These contain more variable similarity clusters to the Arabidopsis sequences than the synthetic FAD2 fragments and showed cross hybridization thresholds between 60% to 70%. Based on these experiments it is clear that some closely related gene family members will not be discriminated. However, with complete availability of the Arabidopsis genome it is possible to assess the approximate extent of potential cross hybridization. For example, most of the seven known Arabidopsis acyl carrier protein (ACP) genes are less than 70% identical and unlikely to cross hybridize, whereas four of the five members of the stearoyl-ACP desaturase family are >80% identical (Mehkedov et al., 2000). As shown in Figure 1 and as described in “Materials and Methods,” additional controls monitored for nonspecific hybridization carry over during printing and for mRNA integrity/probe length.
Figure 2.
Detection of cross hybridization. The fluorescence intensity values of four different FAD2 fragments are plotted for three cohybridization experiments with Cy3/Cy5 probes. The corresponding tissues and fluorescence dyes used for probe synthesis are given on the right. The four FAD2 fragments are displayed on the x axis in the following order: 1,100-bp form of FAD2, 365-bp fragment with 100% identity to the 3′-coding area of FAD2, and two synthetic fragments with 90% and 80% sequence identity, respectively, covering the same 3′-segment of FAD2 as the 100% fragment. All four sequences have identical GC content (48%). The two synthetic fragments contain evenly spaced mismatches and were synthesized by a PCR strategy with four overlapping 110-mer primers (displayed in box in upper right corner).
Microarray Hybridizations
To monitor seed-specific gene expressions, mRNA samples from seeds, leaves, and roots of Arabidopsis were isolated and reverse transcribed with oligo-dT primers into first-strand cDNA fluorescent probes. The mRNA isolated from seeds was the reference to which the samples from leaves and roots were compared. Each tissue comparison was performed at least twice using, in most cases, independently isolated RNA samples as starting material. For repeated experiments the probe pairs contained the fluorochromes Cy3 and Cy5 in opposite orientation. Results of repeated experiments were only used for further analyses if the ratios of all data points on the array showed a correlation coefficient close to one. To eliminate highly variable and therefore less reliable expression data we used data for further analysis only if at least two experiments showed the same trend of expression. Averaging ratios across experiments was considered a less stringent strategy because it neglects the variability between measurements (DeRisi et al., 1997). This is particularly true when low tissue mass (as with developing Arabidopsis seeds) is a limitation for the number of feasible experiments. For the experiments described here, over 20 h of dissection of developing seeds from siliques was required to harvest material for a single fluorescent probe.
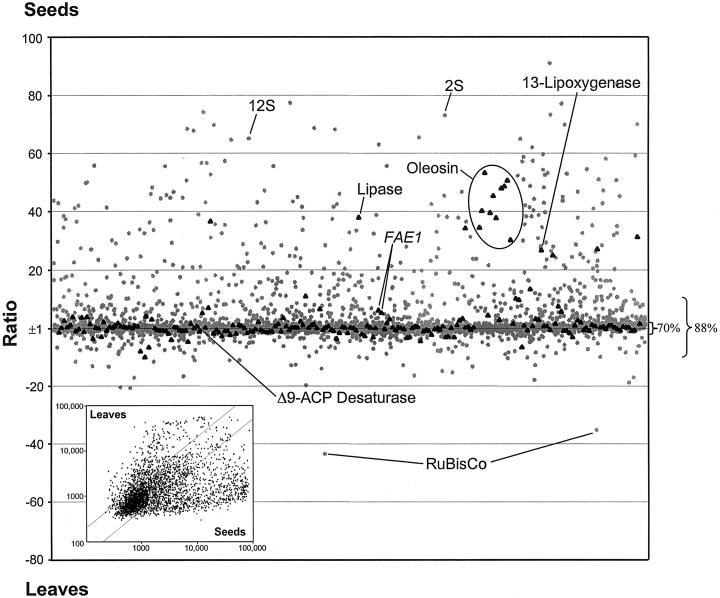
A scatter plot of the data for a seed versus leaf comparisons is shown in Figure 3. It is clear from this representation that the majority of genes analyzed fall near the_x_ axis and have less than a 2-fold difference in signal intensity between the leaf and seed probes. Thus, although the microarray was based on a set of ESTs primarily derived from sequencing of a seed cDNA library, the overall expression pattern shown in Figure3 clearly indicates that a large proportion of seed expressed genes are also expressed in other tissues. These data support the general conclusion based on hybridization analysis of RNA complexity that 60% to 77% (the majority) of plant genes do not have strong tissue-specific expression (Kamalay and Goldberg, 1980; Okamuro and Goldberg, 1989). Expression analyses with smaller and non-seed specific arrays from Arabidopsis detected comparable amounts of tissue specific (Ruan et al., 1998) or differentially expressed genes (Desprez et al., 1998; Kehoe et al., 1999; Richmond and Somerville, 2000).
Figure 3.
Scatter plot of the ratios of the normalized fluorescence intensity values from a seed versus leaf comparison. Expression values that are higher in seeds are plotted upwards and those that are higher in leaves are plotted downwards. Ratios from sequences involved in lipid metabolism and related pathways are displayed with black triangles. All other sequences are represented with gray dots. The percentage values on the right side represent the amount of signals localized within the ratio ranges of ±2 and ±10. Inset, Plot of leaf intensity values versus seed intensity values from same data set.
Nevertheless the microarrays reveal that a substantial number of genes can be considered seed-specific. In the seed versus leaf cohybridizations, approximately 30% of the spotted cDNAs showed more than 2-fold stronger signals in seeds and approximately 12% were expressed more than 10-fold higher in seeds than in leaves (TableI). In the corresponding seed versus root experiments similar comparisons yielded 33% and 13% of the genes, respectively. If both tissue comparisons are combined, 25% of genes showed more than 2-fold and 10% more than 10-fold stronger signals in seeds than in leaves or roots. One factor should be noted that influences these numbers. The reliability of the signals used to calculate these ratios was ensured by including only those values that showed fluorescent intensity levels in at least one channel above three times the local background. This high signal-to-noise ratio and the stringent limit for the ratios of more than 2-fold in each experiment of both tissue comparisons selects preferentially for genes that are moderate to strongly expressed in seeds and only to a very low extent in the other tissues. A disadvantage of this sorting for high confidence values is its tendency to disregard weakly expressed genes, which generally do not reach a high and stable enough signal-to-background ratio in several experiments to appear in this list.
Table I.
Number of genes with seed-specific expression patterns
| Expression Ratioa | Seed/Leaf | Seed/Root | Seed/Leaf and Root |
|---|---|---|---|
| ≥2 | 804 (30%)b | 899 (33%) | 688 (25%) |
| ≥4 | 555 (21%) | 615 (23%) | 478 (18%) |
| ≥10 | 325 (12%) | 348 (13%) | 264 (10%) |
Characteristics of the Seed-Expressed Set
The tissue-expression ratios for a number of well-characterized genes and the variability observed in replicated experiments is presented in Table II. The set of highly seed-specific expressed sequences (ratio ≥ 4) contains several seed storage proteins and a number of other genes that are well known to be predominantly seed expressed. These include oleosins (Abell et al., 1997), fatty acid elongase (FAE1;James et al., 1995), lipoxygenase (Fauconnier et al., 1995), and other genes. In a similar manner, our arrays included a number of genes involved in photosynthesis and carbon fixation such as chlorophyll_a/b_-binding protein and the small subunit of Rubisco. These and other related photosynthetic genes were found to be expressed preferentially in leaves. Thus the overall reliability of the microarrays was confirmed by obtaining the expected preferential seed or leaf expression patterns for dozens of well-characterized genes.
Table II.
Selected examples of Arabidopsis and oilseed rape expression patterns
| Putative Identitya | Clone ID | Ratiob | ||||
|---|---|---|---|---|---|---|
| Seed/leaf | Seed/root | |||||
| B.n.c | A.t.1d | A.t.2 | A.t.1 | A.t.2 | ||
| Storage proteins | ||||||
| 12S Storage protein | M29F06 | 25 | 66 | 92 | 27 | 21 |
| 2S Storage protein 4 | M15F11 | 16 | 28 | 62 | 13 | 18 |
| Lipid metabolism | ||||||
| Biotin carboxyl carrier protein (oilseed rape) | M53F09 | 2 | 5 | 8 | 4 | 4 |
| ω-3 Fatty acid desaturase (endoplasmic reticulum) | M43E07 | 7 | 4 | 3 | 4 | 3 |
| Fatty acid elongase (FAE1) | M16H06 | 16 | 6 | 8 | 8 | 7 |
| Diacylglycerol acyltransferase | E6B2T7 | 2 | −2 | 3 | 2 | −2 |
| Lipoxygenase (Solanum tuberosum) | M30E03 | 6 | 14 | 19 | 18 | 12 |
| Lipid transfer protein 2 | M44A11 | 2 | 11 | 11 | 8 | 10 |
| Oleosin | M36H07 | 41 | 55 | 59 | 35 | 32 |
| Carbohydrate metabolism | ||||||
| Pyruvate dehydrogenase E1 alpha subunit | M11E10 | 13 | 8 | 12 | 8 | 10 |
| Transcriptional regulation | ||||||
| Pc-Myb2-like protein | M18D02 | 3 | 11 | 25 | 16 | 15 |
| Zinc finger protein | M18H11 | 7 | 17 | 23 | 18 | 12 |
| CCAAT-box binding factor HAP3 | M38G03 | 2 | 4 | 6 | 6 | 5 |
| Signal transduction | ||||||
| grr1 Homolog (Glycine max) | M41C07 | 12 | 15 | 16 | 14 | 13 |
| Calmodulin-related protein 2 | M12B04 | 8 | 42 | 50 | 24 | 21 |
| Receptor protein kinase homolog | M19D10 | 12 | 19 | 38 | 31 | 14 |
| Putative protein Ser/Thr kinase | M34C01 | 3 | 12 | 14 | 10 | 8 |
| Putative ARF1 GTPase-activating protein | M39G12 | 3 | 2 | 2 | 16 | 9 |
| Development | ||||||
| Endosperm-specific protein (Zea mays) | M60F09 | 1 | 4 | 2 | 3 | 3 |
| Embryo-specific protein 1 | M58H11 | 24 | 13 | 23 | 28 | 16 |
| Embryo-specific protein 3 | M49A09 | 3 | 28 | 24 | 22 | 16 |
| Terminal flower 1 | M36F04 | 5 | 17 | 37 | 16 | 17 |
| Dessication-related protein (Craterostigma plantagineum) | M12D06 | 3 | 22 | 37 | 28 | 12 |
| Photosynthesis | ||||||
| RuBisCo SSU | M54F03 | −18 | −8 | −7 | 11 | 18 |
| Chla/b-binding protein | M18A12 | −5 | −5 | −4 | 17 | 12 |
We previously classified the seed-expressed ESTs according to codes that categorize their putative function (White et al., 2000). TableIII presents a partial summary of the microarray analysis of groups of clones from several categories. Only storage proteins stand out as a class with a high proportion of seed-specific sequences. As observed for the overall set of 2,600 genes (Table I), only a minority of the clones in all other clone categories are seed-specific. Although oil is the major storage reserve in Arabidopsis seeds, lipid biosynthesis-related genes were in general only slightly more highly expressed in seeds. Of the 113 genes included on the microarrays that are related to lipid biosynthesis, only 10 were found to occur in the subset with ≥10-fold higher seed versus leaf or root signals. These numbers reflect the fact that lipid biosynthesis is essential for growth of all tissues and can be considered a “housekeeping” function. The 10 lipid-related genes with high seed-to-leaf/root expression ratios include oleosin, FAE1, and lipases.
Table III.
Summary characteristics of seed-specific genes
| Code | Code Description | ≥2 | ≥4 | ≥10 | Percentage of Class ≥2a |
|---|---|---|---|---|---|
| AA | Amino acid metabolism | 3 | 3 | 3 | 5 |
| C | Carbohydrate metabolism | 13 | 11 | 6 | 9 |
| DEV | Development | 19 | 14 | 10 | 25 |
| DNA | DNA-modifying enzymes | 2 | 1 | 1 | 7 |
| L | Lipid metabolism | 23 | 15 | 10 | 20 |
| RB | Ribosome, protein translation | 5 | 3 | 1 | 6 |
| RNA | Acting on RNA | 5 | 4 | 3 | 11 |
| SEM | Secondary metabolism | 7 | 5 | 3 | 12 |
| SP | Storage protein | 6 | 6 | 6 | 86 |
| STD | Signal transduction (kinases, etc.) | 19 | 11 | 3 | 14 |
| T | Transcription factors | 8 | 3 | 2 | 12 |
| UF | Unidentified function | 56 | 37 | 20 | 13 |
| NSH | Nonsignificant homology | 112 | 74 | 37 | 13 |
An approximate 28 cDNAs with homology to transcription factors, kinases, phosphatases, and proteins involved in development were highly seed-specific (ratio ≥ 4). Most of these represent genes that have not previously been characterized at the level of tissue-specific expression. Over 110 cDNAs of the ≥4-fold subset (more than 23%) show no significant homology to known sequences (BLAST score <100) or fall in the category of proteins with unidentified function. Because the sequences of most structural genes are known, it is likely that these sets of new and unidentified seed-specific sequences contain many additional regulatory genes.
Identification of New Strong Seed-Specific Promoters
Because EST abundance is in most cases related to mRNA abundance, the sequencing of >10,000 ESTs from a seed cDNA library has provided a set of data that can be used to identify highly expressed genes (White et al., 2000). Microarray data as described here provides additional information on tissue specificity of gene expression. By combining these two types of data, it is possible to identify genes that are strongly expressed and expressed with high tissue specificity. Of course many seed storage proteins and other genes are well known to fall into this category. In Table IV we have identified a number of additional such candidates that have high EST abundance and high seed specificity based on microarrays. Many of these highly expressed genes encode proteins of unidentified function and therefore may be of particular interest in future functional genomic studies of seed metabolism and development. In addition, the promoters from such genes may be useful to control the expression of economic traits in the production of transgenic plants and further examination may reveal that some have particularly useful timing of expression during embryogenesis.
Table IV.
Examples of highly expressed and seed-specific genes
| Putative Identitya | Clone ID | Average | Contigsb | ||
|---|---|---|---|---|---|
| Seedc | Leafc | Ratiod | No. of ESTs | ||
| Dessication protein (C. plantagineum) | M12D06 | 12,602 | 447 | 30 | 59 |
| Germin-like protein (GLP2a) | M31F10 | 34,810 | 1,005 | 43 | 11 |
| Unknown protein | M42C12 | 39,349 | 1,338 | 37 | 18 |
| Unknown protein | M20H04 | 44,624 | 1,895 | 30 | 18 |
| Unknown gene | M36D01 | 64,744 | 999 | 70 | 17 |
| Unknown gene | M42A08 | 27,195 | 4,104 | 7 | 15 |
| Hypothetical protein | M40D09 | 67,359 | 10,059 | 9 | 12 |
| Putative high-pI laccase (Oryza sativa) | M18A04 | 4,009 | 380 | 12 | 22 |
Application of Arabidopsis Microarrays to Oilseed Rape
Species within the genus Brassica are the major vegetable oil crop grown in northern Europe, Canada, and China and represent the third largest source of vegetable oils worldwide. Because of the close phylogenetic relationship of Arabidopsis to_Brassica_ we examined the ability of the arrays developed for this study to provide information on gene expression in oilseed rape. When hybridized with seed and leaf mRNA samples, the correlation coefficients between Arabidopsis and Brassica experiments varied between 0.73 and 0.83 for ratios and 0.76 and 0.83 for intensities (Table V). Because these values are only slightly lower than those for repeated Arabidopsis experiments, which varied between 0.86 and 0.87 for ratios and 0.84 and 0.96 for intensities, it is clear that Arabidopsis microarrays are a very useful tool to analyze related Brassica species. In addition, most seed-specific sequences, which we identified here with Arabidopsis probes (Table I and website), also gave seed-specific signals in the Brassica hybridization. However, the averaged signal intensities of Brassica experiments are approximately 2-fold lower than those from Arabidopsis experiments and, although 80% of genes gave signals at least 2-fold over background with Arabidopsis probes, this number was reduced to 50% with oilseed rape. Therefore, the signals from some weakly expressed genes are likely to be lost in experiments with heterologous probes.
Table V.
Correlation between experiments
| Experiment | Ata Seed/Root 2 | At Seed/Leaf 2 | Bnb Seed/Leaf |
|---|---|---|---|
| At Seed/Root 1 | |||
| Ratio | 0.87 | ||
| Int (seed)c | 0.96 | 0.96 | 0.83 |
| Int (root) | 0.84 | ||
| Int (seed/root) | 0.17 | ||
| At Seed/Leaf 1 | |||
| Ratio | 0.86 | 0.83 | |
| Int (seed) | 0.89 | 0.92 | 0.83 |
| Int (leaf) | 0.91 | 0.77 | |
| Int (seed/leaf)d | 0.12 | 0.16 | |
| Bn Seed/Leaf | |||
| Ratio | 0.73 | ||
| Int (seed) | 0.79 | 0.80 | |
| Int (leaf) | 0.76 | ||
| Int (seed/leaf) | 0.20 |
DISCUSSION
The data set derived from this study provides initial characterization of the tissue expression patterns for a large number of Arabidopsis genes. For a substantial number (at least 2,000) of the genes studied here no previously published data are available on their expression patterns in seeds or other tissues and therefore these data provide initial information useful toward their characterization. Furthermore, at least 40% of the genes on the arrays are of unknown function, and therefore these new data can guide future work in functional genomics. As just one example, knowledge that previously uncharacterized protein kinases (such as clones M19D10 and M34C01) are seed-specific can direct future analysis of the phenotype of mutants or transgenic plants altered in their expression. In a similar manner, a number of uncharacterized transcription factors are defined by these data as having seed-specific expression and further analysis of their function may provide clues regarding transcriptional control of seed metabolism and development.
The data set also defines a large number of seed-specific genes that can be further analyzed by examination of the promoter regions for these genes. Only a handful of genes have previously been available for such analysis , which included primarily seed storage protein or other genes with highly abundant transcripts. The set described here includes a much wider range of examples, including genes with widely different expression levels. Bioinformatics analysis of several hundred such promoters with approaches similar to those described by Hughes et al. (2000), Tavazoie et al. (1999), or Zhang (1999) may therefore offer new insights on cis activation sequences responsible for control of seed expression. Moreover, these promoters can be used to clone their corresponding trans acting elements using yeast one-hybrid screenings or similar approaches.
Several crop plants are phylogenetically close to Arabidopsis and we therefore explored the ability of Arabidopsis based arrays to provide useful information on such species. When hybridized with probes derived from mRNA isolated from oilseed rape, the Arabidopsis arrays provided a very useful data set with only a minor loss in sensitivity. The microarray technique thus will enable detailed studies of gene expression in different Brassica cultivars. We are currently using the arrays to analyze seeds from transgenic oilseed rape lines. These results furthermore suggest that other species within the Brassicaceae (e.g. broccoli, cabbage, mustards, etc.) can likely be analyzed with Arabidopsis based arrays. This ability is a feature of the cDNA/glass slide-based arrays used here that will continue to make them attractive alternatives to oligonucleotide based arrays (Lipshutz et al., 1999) for analysis of many species. The possibility to analyze related species with the same microarray also makes it feasible to compare Arabidopsis and Brassica mRNA populations directly by simultaneous hybridization of mixed probes to the same microarray. It is intriguing that in preliminary experiments with such heterologous comparisons, a number of genes are clearly expressed more highly in the heterologous (oilseed rape) sample.
Limitations to Microarray Analysis
Based on spiking of our mRNA preparations with internal standards, it can be estimated that the sensitivity of these microarrays is approximately one mRNA species per 100,000. This roughly corresponds to one to two mRNA molecules per cell based on the estimate that a cotton embryo cell contains approximately 120,000 mRNA molecules per cell (Dure et al., 1981; Galau and Dure, 1981). This level of sensitivity is thus sufficient to detect a large proportion of all genes expressed in the developing seeds. However, it should be recognized that there are other factors that limit the amount of data obtainable from these arrays. Most importantly, the arrays that we have produced, although containing thousands of genes, currently do not contain a high representation of rarely expressed genes. Because the arrays in this initial study are based on sequencing the first 5,000 of 10,500 ESTs from a partially subtracted cDNA library, mRNAs of abundance lower than 0.01% will be under-represented in the population of genes surveyed by these microarrays. Future generations of microarrays that include much more complete coverage of the Arabidopsis genome will become available and allow extension of the current data. However, it should be recognized that current microarray technology, whether cDNA- or oligonucleotide-based, will continue to have difficulty in reliable detection of the most rarely expressed genes. The presence of many highly abundant transcripts, as those for seed storage proteins, has a dilution effect on low abundant transcripts. Furthermore, the use of complex tissue samples for probe synthesis consisting of different and non-synchronized cell types causes an additional increase in probe complexity and can prevent the detection of transcripts that are only expressed in a small proportion of the tissue sample. Laser capture systems for collecting specific cell types and subsequent RNA amplification methods, used with animal cells (Luo et al., 1999), may circumvent some of these limitations specific to microarray analysis of multicellular organisms.
A further limitation to wide-scale transcription profiling based on cDNA arrays is the possibility of cross-contamination of DNA samples. Handling of many thousands of samples in high-density microtiter format through many steps of manipulations introduces the possibilities of cross-contamination via aerosols or other processes. If a 0.1% contamination were to occur between a seed storage protein that is expressed as 1% of the mRNA population and a transcription factor clone that is expressed at 0.001% then the expression profile observed for the transcription factor could artifactually appear as highly seed specific. Such artifacts cannot be detected by resequencing of the clones used to spot the arrays or by many other common controls. Although the great majority of the data from a microarray are valid, this example emphasizes that users of microarray data must always consider the data to be preliminary and require independent confirmation by techniques such as northern analysis.
CONCLUSIONS
The microarrays described in this study have already provided new data on the expression profiles of over 2,000 Arabidopsis genes. A more complete set of data from this study than can be provided here is downloadable at our website (http://www.bpp.msu.edu/Seed/SeedArray.htm) and undoubtedly other workers will be able to “mine” further useful insights by asking questions not considered here. This type of data represents a survey of transcription profiles and is best cataloged in central databases where it can be linked to other types of information as these accumulate for each gene. Therefore, in addition to the web database for this project, data will be available through The Arabidopsis Information Resource (www.Arabidopsis.org) as software to accommodate it is developed. It is clear that the present study provides only the initial information that can be derived from such microarrays. A second stage of more focused analysis will develop in the future where detailed studies of the timing of gene expression and patterns of gene expression in seed mutants such as wri1 (Focks and Benning, 1998) and in transgenic plants will provide a second generation of rich information useful for understanding the complexities of seed metabolism and its control.
MATERIALS AND METHODS
Amplification of cDNAs
The plasmids of 2,715 selected cDNA clones were collected from a cDNA library of developing Arabidopsis seeds. All sequences have been deposited in GenBank dbEST database and are described further in White et al. (2000). An additional 82 cDNA clones from genes of lipid and carbohydrate metabolism were supplied by T. Newman (Michigan State University) and other colleagues. The inserts of the cDNAs were amplified by PCR in a 96-well format using primer pairs specific for the vector ends (for inserts in pBluescript SK−: T7, 5′-GTAATACGACTCACTATAGGGC, and 5′ extended M13 reverse, 5′-ACAGGAAACAGCTATGACCATG; for inserts in pZipLox1: M13 forward, 5′-CCCAGTCACGACGTTGTAAAACG, and M13 reverse, 5′-AGCGGATAACAATTTCACACAGG). PCR reactions of 100-μL volume contained 0.4 μm of each primer, 0.2 μm of each desoxynucleotide, 10 mm Tris [tris(hydroxymethyl)aminomethane], 50 mm KCl, 3.0 mm MgCl2, 3 units of Taq DNA polymerase (Promega, Madison, WI), and approximately 10 ng of plasmid template. The reactions were run on a 9700 Thermoblock (Perkin-Elmer, Foster City, CA) using an amplification program of 3 min denaturation at 94°C, 5 precycles of 30 s at 94°C, 30 s at 64°C, and 2 min at 72°C, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 2 min at 72°C, and terminated by a 7-min extension at 72°C. The PCR products were precipitated by adding 200 μL of ethanol (95%, w/v) and 10 μL of sodium acetate (3m, pH 5.2) and centrifugation at 3,200_g_ at 4°C for 60 min. After washing with 80% (w/v) ethanol, the DNA was resuspended in 20 μL of 3× SSC. The yield and purity of the PCR products was analyzed by agarose gel electrophoresis. PCR samples showing, by agarose gel analysis, concentrations less than 0.2 μg/μL and/or double bands were repeated. If possible, alternative clones from the cDNA clone collection were used to repeat the PCR experiments. To reduce the cross-contamination risk in the 96-well format, failed PCRs were not removed from the sample set and as a result, the number of PCR samples for printing increased by approximately 20%.
Preparation of the cDNA Microarrays
Microscope slides (Gold Seal, No. 3010) were cleaned for 2 h in alkaline washing solution (25 g NaOH in 100 mL of water and 150 mL of 95% [w/v] ethanol), washed in distilled water (five times, 5 min), and then coated for 1 h in 250 mL of coating solution (25 mL of poly-L-Lys, [Sigma, St. Louis], 25 mL of sterile filtered phosphate-buffered saline, and 200 mL of water). After coating, the slides were rinsed with water, dried by centrifugation (5 min at 600 rpm), and subjected to 10 min at 45°C in a vacuum oven. After coating, the slides were cured in a slide box for at least 2 weeks.
PCR samples were arrayed in duplicates from 384-well plates with a center-to-center spacing of 260 μm onto poly-L-Lys-coated slides using a printing device (GeneMachines, San Carlos, CA) with 16 titanium pins (TeleChem, Sunnyvale, CA). The resulting arrays contained 7,680 elements with a size of 18 × 36 mm. After printing, the arrays were rehydrated over a water bath (50°C–60°C) for 15 s, snap-dried for 5 s on a heating block (80°C), and UV crosslinked with a UV 1800 Stratalinker (Stratagene, La Jolla, CA) at 65 mJ of energy. After crosslinking, the remaining functional groups of the surface were blocked for 15 min in blocking solution (4.28 g succinic anhydride [Aldrich, Milwaukee, WI], dissolved in 239 mL of 1,2-methyl-pyrrolidinone (Aldrich), and 10.7 mL of 1 m boric acid, pH 8.0, with NaOH). After blocking, the bound DNA was denatured for 2 min in distilled water at 95°C, rinsed with 95% (w/v) ethanol at room temperature, and finally dried by centrifugation (5 min at 600 rpm).
Controls
To monitor the detection sensitivity limit, the inserts of nine human cDNA clones (IMAGE Ids: 1593326, 1420858, 1484059, 978938, 1593605, 1020153, 1592600, 1576490, and 204625) were amplified by PCR and arrayed at four different locations of the slide. Before probe synthesis, the corresponding mRNA species in vitro transcribed from these human clones were added as internal standards to 1 μg of the plant mRNA samples at levels from 1.0 × 10−3 to 1.0 × 10−5 ng.
To evaluate the hybridizations specificity, a 365-bp long PCR fragment from a FAD2 cDNA clone (L26296) and two synthetic fragments with 90% and 80% sequence identity to the_FAD2_ fragment were arrayed adjacent to each other. The related fragments were synthesized by PCR using four overlapping 110-mer primers into which the required nucleotide exchanges were introduced (Dillon and Rosen, 1990; De Rocher et al., 1998). The resulting three fragments were of equal length and constant GC content. Two additional specificity control sets with more variable similarity clusters in their sequence were spotted as well. These sets contained ferredoxin cDNA sequences from Arabidopsis, Anabaena (M14737), Thunbergia, soybean, Impatiens (supplied from D. Schultz), and for ACP-desaturases from Arabidopsis (M40E01),Geranium (U40344 and AF020203), and Coriandrum sativum (M93115). Unspecific background hybridizations were monitored with PCR products from 12 human cDNAs (IMAGE IDs: h29512, h00641, t91128, 680973, 237257, 280523, 136643, 204716, 60027, 756944, 29328, and IB187) arrayed in several copies at various locations of the array. To analyze the efficiency of the probe synthesis, the 5′-, central, and 3′-regions of two cDNA clones were spotted separately (FAD2, L26296, and a clone for the E1 subunit of the pyruvate dehydrogenase, M20C09). Constant signal intensities of these spots indicated that the probe synthesis by reverse transcription resulted in sufficient amounts of long products. The amount of rRNA contaminations in the hybridization probes were measured with DNA sequences coding for 25S rRNA and 18S rRNA from Arabidopsis. Unspecific probe binding mediated by the poly(A) tail of the cDNAs was detected with arrayed poly(A)50 oligos. The washing efficiency of the spotting pins during the printing process was analyzed by arraying a sequence for Rubisco SSU (118D13T7) and a negative control containing only 3× SSC after each other at several locations of the microarray. To localize the printing grid during the image analysis, the cDNA of a highly expressed translation elongation factor EF-1α (M16D02) was arrayed at two edges of several subgrids.
Plant Material, RNA Extraction, and Probe Synthesis
Arabidopsis ecotype Columbia-2 was grown in a growth chamber with 16 h of light at 80 to 100 μE and temperatures of 22°C during the day and 20°C at night. Developing seeds from each plant type were dissected from siliques at 8 to 11 d after flowering and bulked. Leaf material was collected from the same plants of the same age. Total root tissue was collected from plants grown for 6 weeks in sealed tissue culture boxes containing 50 mL of growth media (1× Murashige and Skoog salts, 1× B vitamins, and 0.5% [w/v] agarose). Oilseed rape (Brassica napus cv 212/86, line 18) was grown in a green house (Eccleston and Ohlrogge, 1998). Seeds were collected from oilseed rape siliques 25 to 30 d after flowering and leaves were collected from the same plants of the same age.
Total RNA was extracted from 1.0 g of plant tissue as described bySchultz et al. (1994). The quality of each total RNA sample was confirmed in a reverse transcription (Superscript II, Boerhinger Mannheim, Basel) test reaction in the presence of [32P]dATP following the manufacturer's instructions. The labeled single-stranded DNA products were separated by agarose gel electrophoresis. The gel was dried and then labeled products were visualized for 1 h using autoradiography. Only RNA samples producing sufficient product in this test labeling were used for subsequent fluorescent probe synthesis. Poly(A)+ RNA was isolated from 100 μg of total RNA using Oligotex oligo(dT) beads (Qiagen, Valencia, CA) following the manufacturer's instructions. Preparation of fluorescent DNA probe was performed as follows: 1 μg of poly(A)+ RNA was mixed with 4 μg of oligo(dT) primer and 1 ng of internal standard in a final volume of 26 μL. This mixture was incubated at 68°C for 10 min, chilled on ice, and then added to 24 μL of reaction mix with a final composition of 1× Superscript II buffer; 500 μm each of dATP, dTTP, and dGTP; 200 μm dCTP; 60 μm Cy3 or Cy5-dCTP (Amersham Pharmacia, Piscataway, NJ); 10 mm dithiothreitol; 1 μL of RNAsin (Boehringer Mannheim); and 3 μL of Superscript II (600 units, Life Technologies, Rockville, MD). The reaction was incubated at 42°C for 60 min, then an additional 360 units of Superscript II was added and incubation was continued at 42°C for another 60 min. After addition of 10 μL of 1 n NaOH, incubation was continued at 37°C for 60 min. 1 m Tris-HCl (25 μL, pH 7.5) was then added and the reaction mix was diluted with 915 μL of Tris-EDTA buffer, followed by extraction with 1 vol of phenol:chloroform (1:1, v/v) and then 1 vol of chloroform:isoamylalcohol (24:1, v/v). The labeled cDNA products were finally transferred to a Centricon 30 filtration column (Millipore, Bedford, MA), washed twice with 2 mL of Tris-EDTA buffer, and then concentrated to a final volume of 10 to 15 μL using a speed vac. Prior to this final concentration step, 1/100 of the labeled probe (approximately 2–4 μL) was removed to determine the quality of the labeling reaction by gel electrophoresis, followed by analysis of the fluorescent signal from the separated products using a ScanArray 3000 laser scanner (GSI Lumonics, Watertown, ME).
Hybridization
Probe mixtures in a total volume of 24 μL were mixed with 6 μL of blocking solution (10 μg/μL of yeast tRNA [Sigma] and 10 μg/μL of oligo-dA [Pharmacia]), 6.3 μL of 20× SSC, and 1.2 μL of 10% (w/v) SDS. The solution was denatured for 1 min at 100°C, cooled down to room temperature, and applied to the array. After covering the array with a 24 × 40 mm coverslip, the slide was placed in a humidified hybridization chamber (TeleChem, Sunnyvale, CA). The hybridization was performed in a 64°C water bath for approximately 16 h. After hybridization, the slides were washed in 1× SSC, 0.2% (w/v) SDS for 5 min, then in 0.1× SSC, 0.2% (w/v) SDS for 5 min, and finally in 0.1× SSC for 30 s. Following the last washing, the slides were immediately dried by centrifugation (5 min at 600 rpm).
Analysis and Quantification
Hybridized microarrays were scanned sequentially for Cy3- and Cy5-labeled probes with a ScanArray 3000 laser scanner at a resolution of 10 μm. To maximize the dynamic range of each scan without saturating the photomultiplier tube and to balance the signal intensities of the two channels approximately, laser power and photomultiplier tube settings of the instrument were adjusted according to the “Auto-Range” and “Auto-Balance” features of the instrument. Signal quantification was performed with the ScanAlyze 2.21 software written by Michael Eisen (available on the Internet:http://rana.stanford.edu/software). The two intensity values of duplicated DNA spots were averaged and used to calculate the intensity ratios between the two channels. Ratios below 1.0 were inverted and multiplied by −1 to aid their interpretation. Intensity values below three times their local background were deemed non-significant and excluded from further data analysis. Since subtraction of the local background from the intensity values often results in artificially high ratios, this operation was not performed for calculating the ratios. Normalization of the intensity values from the two channels was performed by stepwise exclusions of 5% of the highest and 5% of the lowest ratios and calculating for the remaining subsets the mean ratios. It was usual that after excluding 15% of the highest and 15% of the lowest values, the calculated mean ratios reached a plateau, which showed only minor changes in the smaller subsets. The average value of the remaining 70% ratios was used to normalize the intensity ratios as close to 1.0 as possible. The accuracy of this filter method was evaluated by comparing it with the normalization factor calculated from the intensity ratios of the human mRNAs spiked into the labeling reaction. In general, the two methods resulted in relatively similar normalization factors. However, since external RNA controls disregard purity and integrity problems of the actual RNA samples, their use for normalization is more error prone than the filter method used for this study.
ACKNOWLEDGMENTS
We thank Tom Newman for supplying Arabidopsis cDNA clones and for help with robotics and Uwe Rossbach for constructing the website. Curt Wilkerson provided advice on data analysis and Kamlesh Shah provided assistance with design of databases. We thank Ellen Wisman for advice and access to microarray equipment.
Footnotes
1
This work was supported in part by the National Science Foundation (grant no. DCB94–06466) and the Consortium for Plant Biotechnology Research. We also acknowledge the Michigan Agricultural Experiment Station for its support of this research.
LITERATURE CITED
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- De Rocher EJ, Vargo-Gogola TC, Diehn SH, Green PJ. Direct evidence for rapid degradation of Bacillus thuringiensis toxin mRNA as a cause of poor expression in plants. Plant Physiol. 1998;117:1445–1461. doi: 10.1104/pp.117.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Amselem J, Caboche M, Hofte H. Differential gene expression in Arabidopsis monitored using cDNA arrays. Plant J. 1998;14:643–652. doi: 10.1046/j.1365-313x.1998.00160.x. [DOI] [PubMed] [Google Scholar]
- Dillon PJ, Rosen CA. A rapid method for the construction of synthetic genes using the polymerase chain reaction. Biotechniques. 1990;9:298–300. [PubMed] [Google Scholar]
- Dure L, 3d, Greenway SC, Galau GA. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry. 1981;20:4162–4168. doi: 10.1021/bi00517a033. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S. Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 2000;122:767–774. doi: 10.1104/pp.122.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston VS, Ohlrogge JB. Expression of lauroyl-acyl carrier protein thioesterase in Brassica napus seeds induces pathways for both fatty acid oxidation and biosynthesis and implies a set point for triacylglycerol accumulation. Plant Cell. 1998;10:613–622. doi: 10.1105/tpc.10.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauconnier ML, Vanzeveren E, Marlier M, Lognay G, Wathelet JP, Severin M. Assessment of lipoxygen-ase activity in seed extracts from 35 plant species. Grasas Aceites. 1995;46:6–10. [Google Scholar]
- Focks N, Benning C. wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118:91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann LH, Yoon ES, Meinke DW. Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J. 1995;7:341–350. [Google Scholar]
- Galau GA, Dure L., III Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by reciprocal heterologous complementary deoxyribonucleic acid-messenger ribonucleic acid hybridization. Biochemistry. 1981;20:4169–4178. doi: 10.1021/bi00517a034. [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- James DW, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalay JC, Goldberg RB. Regulation of structural gene expression in tobacco. Cell. 1980;19:935–946. doi: 10.1016/0092-8674(80)90085-9. [DOI] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L. Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 1995;108:399–409. doi: 10.1104/pp.108.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe DM, Villand P, Somerville S. DNA microarrays for studies of higher plants and other photosynthetic organisms. Trends Plant Sci. 1999;4:38–41. doi: 10.1016/s1360-1385(98)01354-5. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- Mehkedov S, Martínez de Ilárduya O, Ohlrogge J. Toward a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–402. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Jaworski J. Regulation of plant fatty acid biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Browse J, Somerville CR. The genetics of plant lipids. Biochem Biophys Acta. 1991;1082:1–26. doi: 10.1016/0005-2760(91)90294-r. [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Goldberg RB. Regulation of plant gene expression: general principles. In: Stumpf PK, Conn EE, editors. The Biochemistry of Plants. Vol. 15. New York: Academic Press; 1989. pp. 1–82. [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T, Somerville S. Chasing the dream: plant EST microarrays. Curr Opin Plant Biol. 2000;3:108–116. doi: 10.1016/s1369-5266(99)00049-7. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Gilmore J, Conner T. Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DJ, Craig R, Cox-Foster DL, Mumma RO, Medford J. RNA isolation from recalcitrant plant tissue. Plant Mol Biol Rep. 1994;12:310–316. [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Thomas TL. Gene expression during plant embryogenesis and germination: an overview. Plant Cell. 1993;5:1401–1410. doi: 10.1105/tpc.5.10.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Girke T, Focks N, Martinez de Ilárduya O, Jaworski JG, Ohlrogge J, Benning C. A new set of Arabidopsis ESTs from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ. Promoter analysis of co-regulated genes in the yeast genome. Comput Chem. 1999;23:233–250. doi: 10.1016/s0097-8485(99)00020-0. [DOI] [PubMed] [Google Scholar]