PubChem 2019 update: improved access to chemical data (original) (raw)
Abstract
PubChem (https://pubchem.ncbi.nlm.nih.gov) is a key chemical information resource for the biomedical research community. Substantial improvements were made in the past few years. New data content was added, including spectral information, scientific articles mentioning chemicals, and information for food and agricultural chemicals. PubChem released new web interfaces, such as PubChem Target View page, Sources page, Bioactivity dyad pages and Patent View page. PubChem also released a major update to PubChem Widgets and introduced a new programmatic access interface, called PUG-View. This paper describes these new developments in PubChem.
INTRODUCTION
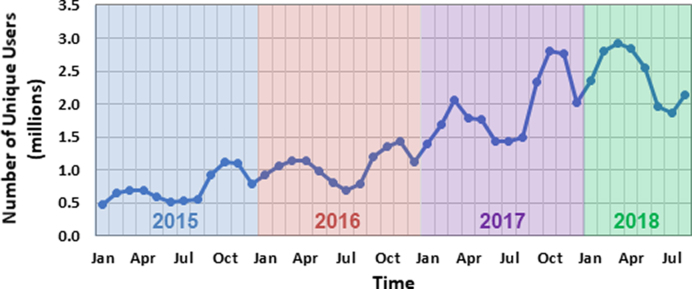
PubChem (https://pubchem.ncbi.nlm.nih.gov) (1–3) is a chemical information resource at the U.S. National Center for Biotechnology Information (NCBI). Since its launch in 2004, PubChem has grown into a key knowledge base that serves the biomedical research communities in many areas, including cheminformatics, chemical biology, medicinal chemistry and drug discovery. PubChem is a popular chemistry web site, with about three million unique interactive users per month at peak usage times (Figure 1).
Figure 1.
Number of unique PubChem users per month (interactive users only).
PubChem organizes its data into three inter-linked databases: Substance, Compound, and BioAssay (1,2). The Substance database archives depositor-contributed descriptions of chemical substances (1). The Compound database stores unique chemical structures extracted from the Substance database through structure standardization (1,4). The BioAssay database contains the description and test results of biological assay experiments (2). An overview of these three databases is given in our previous papers published in the Nucleic Acids Research Database issue (1,2).
The present paper provides an update on the three PubChem databases since the previous papers (1,2). This includes new data content and sources, the introduction of legacy designation for outdated records, and the updated molecular weight values, as well as the release of new web interfaces such as the PubChem Target View page, Bioactivity dyad pages, Patent View page, Sources page and Widgets. Recent changes in PubChem's web services for programmatic access are also summarized in this paper.
DATA CONTENTS
Over the past two years, there has been a significant growth in PubChem data. As of August 2018, PubChem contains 247.3 million substance descriptions, 96.5 million unique chemical structures, contributed by 629 data sources from 40 countries. It also contains 237 million bioactivity test results from 1.25 million biological assays, covering >10 000 target protein sequences.
The spectral information content in PubChem has substantially increased and now >591 000 compounds in PubChem have one or more types of spectral information available, including 13C NMR, 1H NMR, 2D NMR, ATR-IR, FT-IR, MS, GC–MS, Raman, UV–Vis, vapor-phase IR and more. A notable addition of spectral information to PubChem was from SpectraBase (http://spectrabase.com), a free online spectral repository from Bio-Rad Laboratories, Inc. It provided images of, annotations about, and links to a diverse set of spectral information for 225 000 compounds, including NMR, IR, RAMAN, UV and MS. In addition, the MassBank of North America (MoNA; http://mona.fiehnlab.ucdavis.edu) provided meta data for and links to >173 000 mass spectra for 77 000 compounds.
The publisher Springer Nature (https://www.springernature.com) provided PubChem with more than 26 million links between 600 000 chemical substance records and four million scientific articles and book chapters, with updates on a weekly basis. Among these, 1.6 million links point to 300 000 open- or free-access articles. Considering that only ∼20% of the Springer Nature articles could be found in PubMed, this addition enhanced the discoverability and accessibility of chemical information in PubChem, by complementing the existing literature information derived from MeSH annotations to PubMed abstracts (5).
In addition, annotations from authoritative sources for agricultural and food chemicals have been added to PubChem. For example, PubChem now provides information on pesticides, collected from EPA Office of Pesticide Programs (http://www.ipmcenters.org/Ecotox/index.cfm), USDA Pesticide Data Program (https://www.ams.usda.gov/datasets/pdp), and EU Pesticides Database (http://ec.europa.eu/food/plant/pesticides/eu-pesticides-database/public/). Information on food additives and nutrients from FDA Center for Food Safety and Applied Nutrition (CFSAN) (https://www.fda.gov/Food/), EU Food Improvement Agents (https://ec.europa.eu/food/safety/food_improvement_agents/) and Joint FAO/WHO Food Additive Evaluations (JECFA) (http://apps.who.int/food-additives-contaminants-jecfa-database/) have also been integrated within PubChem.
Introduction of legacy designation
As an archive, PubChem accepts scientific data from contributors and maintains that data even if the contributing project is discontinued. Therefore, some records in PubChem can persist with outdated (and, at times, incorrect) data. To help users identify such cases, PubChem introduced a legacy designation for collections that are not regularly updated. This legacy designation applies to collections that no longer appear to be active as well as to their individual records. The PubChem Data Sources page (https://pubchem.ncbi.nlm.nih.gov/sources), explained later in this paper, allows one to figure out who the legacy projects/contributors are. The legacy designation can help PubChem users quickly identify records that may have out-of-date information and/or hyperlinks. More information on the legacy designation is given in a PubChem Help document (https://pubchemdocs.ncbi.nlm.nih.gov/legacy-designation) as well as a PubChem Blog post (http://1.usa.gov/1H52gyW).
Atomic mass changes
Molecular weights in PubChem were updated using the latest International Union of Pure and Applied Chemistry (IUPAC) recommendations for atomic mass and isotopic composition information (6,7). Although the molecular weight computation of a chemical seems straightforward, the scientific community is recognizing complex issues with average atomic weight and isotopic data, as greater degrees of precision in atomic masses are known. Therefore, PubChem now uses conventional atomic weights (6), when available. For the elements without any abundance information (e.g. technetium), the atomic weight of the most stable, non-theoretical isotope was used, as found in NuBase (http://amdc.in2p3.fr/web/nubase_en.html) (8). In addition, PubChem is now restricting the allowed isotopes for a given element to those with a half-life of one millisecond or greater. More detailed information on this change can be found at a PubChem Blog post (http://go.usa.gov/x8RqD).
WEB INTERFACES
PubChem Target View page
PubChem contains 237 million bioactivities for three million compounds, determined in over 1.2 million biological assay experiments. Many of these assays are performed against target proteins or genes. However, finding all bioactivity data for a given target is not a trivial task. The PubChem Target page provides a ‘target-centric’ view of PubChem data pertinent to a given gene or protein target, including the chemicals tested against the target and biological assay experiments performed against the target. The Target View page also presents known drugs and curated ligands, collected from ChEMBL (9), DrugBank (10) and IUPHAR/BPS Guide to PHARMACOLOGY (11). In addition, it contains annotated information about the target, such as synonyms, biological functions, relevance to disease, gene/protein classifications, protein structures, gene-gene interactions, orthologs, pathways, etc. These annotations are collected from major molecular biology databases, including NCBI Gene (12), Gene Ontology (13,14), Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) (15), UniProt (16), Protein Data Bank (PDB) (17), Conserved Domains Database (CDD) (18), Pfam (19), MedGen (20), Kyoto Encyclopedia of Genes and Genomes (KEGG) (21), BioSystems (22), Molecular Modeling Database (MMDB) (23).
The PubChem Target View page for a given target can be accessed via a web unique resource locator (URL) that contains the corresponding NCBI Gene ID or Gene Symbol. For example, the target page for the human histamine receptor H1 (HRH1) gene (NCBI Gene ID: 3269) can be accessed via the URL:
Note that when a gene symbol is used in the URL, the Target View page for the corresponding human gene is presented. The Target View page for a gene from a particular species may be accessed through a URL containing the species name. For example, the following URLs point to the Target View page for the HRH1 gene of the house mouse (mus musculus):
- https://pubchem.ncbi.nlm.nih.gov/target/gene/hrh1/house_mouse
- https://pubchem.ncbi.nlm.nih.gov/target/gene/hrh1/mus_musculus
The target page for a given protein can be accessed via the URL containing the accession number for that protein. For example, the Target View page for the human histamine receptor H1 protein (accession: P35367) can be accessed via the URL:
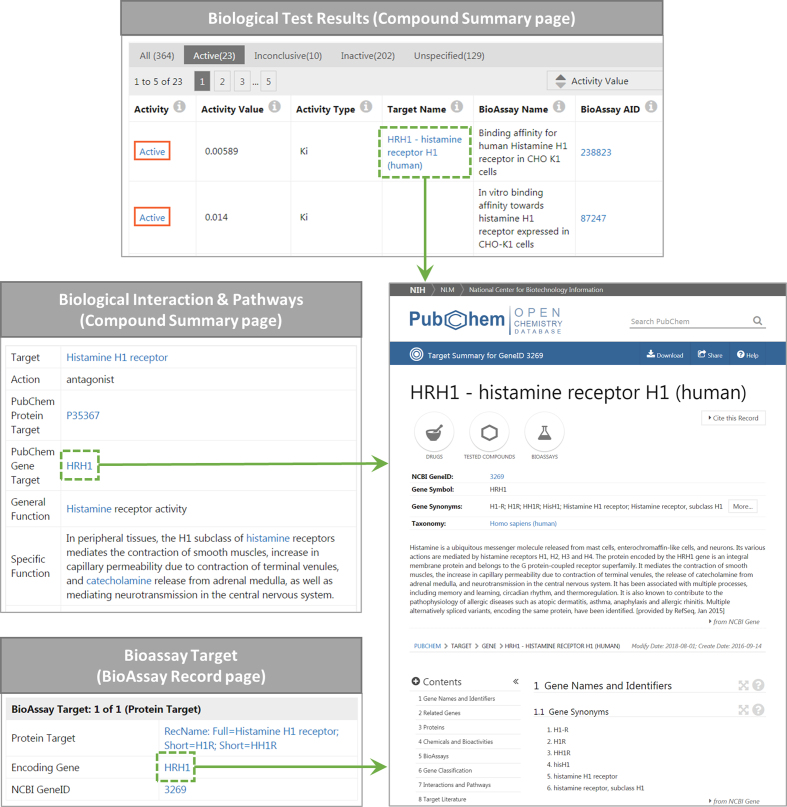
In addition, the Target View page can be accessed from the Summary or Record page of PubChem records (Figure 2). For instance, the Target View page for the human HRH1 gene may be accessed by clicking on the target gene name mentioned in under the ‘BioAssay Results’ or ‘Biomolecular Interactions and Pathways’ section of the Summary page for CID 2678 (Zyrtec). It may also be accessed from the BioAssay Record page of AID 238823.
Figure 2.
PubChem Target View page for the human histamine receptor H1 (HRH1) gene (https://pubchem.ncbi.nlm.nih.gov/target/gene/3269) (bottom right), along with its example entry points from the Compound Summary page for CID 2678 (https://pubchem.ncbi.nlm.nih.gov/compound/2678) and the BioAssay Record page for AID 238823 (https://pubchem.ncbi.nlm.nih.gov/bioassay/238823).
PubChem Bioactivity dyad page
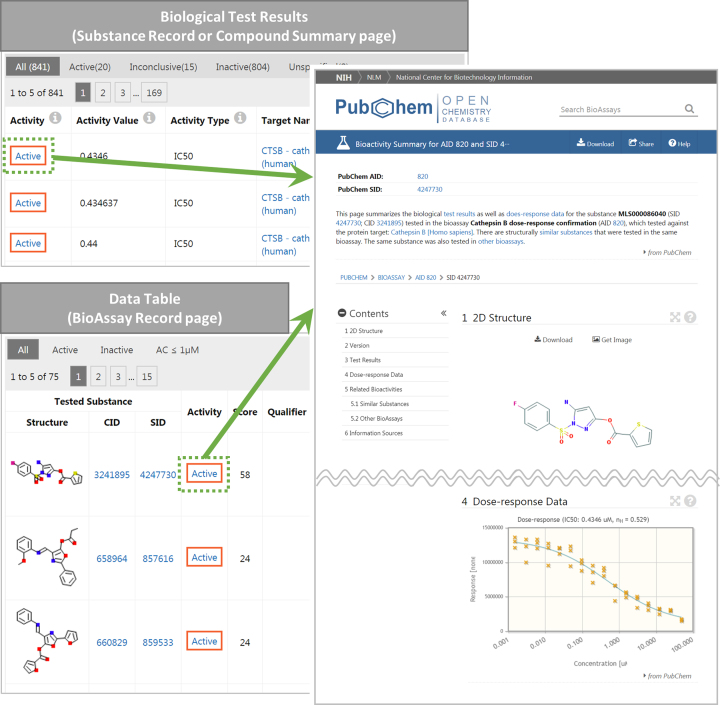
The Bioactivity dyad page presents bioactivity data for a given chemical tested in a particular assay or against a particular gene or protein target. For example, the following AID-SID dyad page presents the bioactivity data of SID 4247730 tested in AID 820.
The dyad page also presents the dose response curve (if available). It also shows the bioactivity data for structurally similar substances tested in the same assay, and the bioactivity data for the same molecule tested in different assays. The AID-SID dyad page may be accessed by clicking the activity outcome (active, inactive, inconclusive or unspecified) displayed in the ‘Activity’ column of the bioassay results table (on the Compound Summary or Substance Record page), or the data table (on the BioAssay Record page), as shown in Figure 3.
Figure 3.
PubChem Bioactivity dyad page for SID 4247730 (corresponding to CID 3241895) and AID 820 (https://pubchem.ncbi.nlm.nih.gov/bioassay/820#sid=4247730) (right). This page can be accessed from the Substance Record page for SID 4247730 (https://pubchem.ncbi.nlm.nih.gov/substance/4247730), the Compound Summary page for CID 3241895 (https://pubchem.ncbi.nlm.nih.gov/compound/3241895), or the BioAssay Record page for AID 820 (https://pubchem.ncbi.nlm.nih.gov/bioassay/238823).
PubChem also provides two additional types of bioactivity dyad pages: the gene-CID and the protein-CID dyad pages, which present the bioactivity data of a given compound tested against a particular gene or protein. For instance, the following URLs are the dyad pages for CID 3241895 and human Cathepsin B gene (Gene ID 1508) and protein (accession P07858).
- Gene-CID dyad: https://pubchem.ncbi.nlm.nih.gov/target/gene/1508#cid=3241895
- Protein-CID dyad: https://pubchem.ncbi.nlm.nih.gov/target/protein/P07858#cid=3241895
The gene-CID and protein-CID dyad pages for a given CID also present the bioactivity data of the same compound against other targets as well as those of structurally similar compounds to that CID against the same target gene or protein. These dyad pages may be accessed from the Target View pages for the corresponding gene and protein target.
Patent View
PubChem contains information on what chemicals are mentioned in patent documents. These chemical-patent associations are generously submitted by several data contributors, including IBM, SureChEMBL (24), NextMove (https://www.nextmovesoftware.com/), SCRIPDB (25), and BindingDB (26). PubChem now organizes this information in a page called the ‘PubChem Patent View’. Below is an example of the patent view page (for U.S. Patent 5969156):
The Patent View page for a given patent provides compounds and substances mentioned in it, along with other information including patent title, abstract, application/publication dates, applicant and inventor. It also contains patent classification information based on World Intellectual Property Organization (WIPO)’s International Patent Classification (IPC).
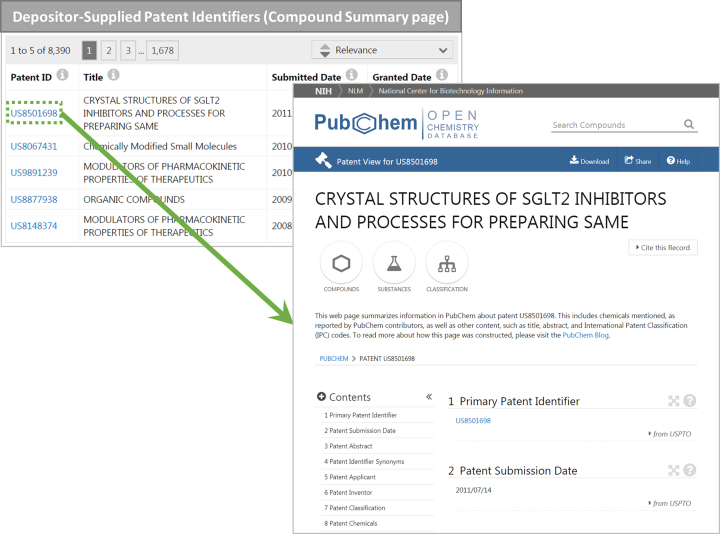
The Patent View page may be accessed by clicking one of patent identifiers listed under the ‘Depositor-Supplied Patent Identifiers’ section on the Compound Summary page (Figure 4). It should be noted that, at the time of writing, the Patent View page does not provide the context about why a particular chemical was mentioned in the patent. In other words, it might not be possible to tell if the chemical is indeed the subject matter of the patent grant, or if it is just mentioned as a part of prior arts in the background section.
Figure 4.
PubChem Patent View page for US8501698 (https://pubchem.ncbi.nlm.nih.gov/patent/US8501698) (right). This page can be accessed from the ‘Depositor-Supplied Patent Identifiers’ section on the Compound Summary page for CID 4247730 (https://pubchem.ncbi.nlm.nih.gov/compound/2162) (left).
PubChem Data Sources page
The PubChem Data Sources page (https://pubchem.ncbi.nlm.nih.gov/sources) is an interface that provides a flexible overview of organizations contributing data to the PubChem. Using PubChem Data Sources page, one can readily find who provided what information to PubChem: substances, assays, and annotations (primarily textual information linked to various types of PubChem records). This interface allows one to filter the data sources by data type, source category, source status, and country, or to sort them by record counts or last-modified date. It is also possible to search a data source by keyword. Clicking a data source name listed on the Data Sources page directs users to a dedicated page for that source, which provides the source URL, contact information, the current counts of records submitted to PubChem, and the date when the content was last updated.
Widget
PubChem has released PubChem Widgets 2.0f (https://pubchemdocs.ncbi.nlm.nih.gov/widgets). PubChem Widgets provide a convenient way for scientific web developers to display PubChem content within webpages they design. Because all data presented in the widgets are served directly from PubChem, the widgets are guaranteed to show most up-to-date content in PubChem. The widgets may be used to display a tabular summary of items linked to PubChem records (e.g. patents, bioactivities, PubMed articles, etc.), a carousel of related chemical structures, or a classification of PubChem record of interest.
PubChem Widgets 2.0f allows one to display any section or subsection of PubChem summary or record pages in a widget (except the top section). Compared to the previous version, the new Widgets provides many more data views and makes them easier to embed into any web page. In addition, the new widgets are easier to resize, which makes them more appropriate for displays with assorted sizes.
PROGRAMMATIC ACCESS
PUG-View
Our previous papers (27,28) describe multiple programmatic access routes to PubChem data, including Entrez Utilities (E-Utilities), Power User Gateway (PUG), PUG-SOAP and PUG-REST (see Table 1). In addition to these existing programmatic interfaces, PubChem introduced a new Representation State Transfer (REST)-style interface, called PUG-View (https://pubchemdocs.ncbi.nlm.nih.gov/pug-view). It was originally developed as a backend service that provides annotations to present on the Summary page for a compound or the Record page of a substance or bioassay, but it can also be used independently as a programmatic web service to access the annotations. For example, the following PUG-View request URL retrieves all annotations presented on the Summary page of CID 2244 (aspirin) in JavaScript Object Notation (JSON) format:
Table 1.
| Access Route | Description |
|---|---|
| Entrez Utilities (E-Utilities) | Suited for accessing text- or numeric-fielded data. No ability to handle complex data types specific to PubChem (e.g. chemical structures, or tabular bioactivity data). |
| Power User Gateway (PUG) | Pure XML-based interface. Uses a complex PubChem-specific XML schema. |
| PUG-SOAP | Uses the Simple-Object Access Protocol (SOAP). Good for scripting/programming languages and automation tools with SOAP interface. |
| PUG-REST | Representational State Transfer (REST)-style interface. Primarily used to access chemical property data computed by PubChem and depositor-provided bioactivity data stored. |
| PUG-View | Representational State Transfer (REST)-style interface. Used to access textual information presented on the PubChem Compound summary or Substance/BioAssay Record page. Used to access data collected from various annotation sources. |
| PubChem RDF REST | Representational State Transfer (REST)-style interface. Used to access RDF-formatted PubChem data. |
It is possible to get particular kinds of annotations for a given PubChem record through PUG-View, by providing the name of the corresponding heading or subheading as an optional parameter after the ‘?’ character. For instance, the following request URL returns the melting point of CID 2244:
Note that the white space in ‘Melting Point’ is replaced with the ‘+’ character.
Some users often confuse PUG-View with another REST-style interface, called PUG-REST, which has been described elsewhere (28). While PUG-REST primarily retrieves property values computed by PubChem, PUG-View retrieves annotations collected from other data sources. A more detailed description of PUG-View is provided at the PUG-View Help page (https://pubchemdocs.ncbi.nlm.nih.gov/pug-view), along with many example request URLs.
Web traffic control through dynamic throttling
While ∼5% of PubChem users are estimated to be programmatic users, PubChem can potentially receive many more programmatic access requests than can be handled, causing unexpected service disruptions. Therefore, PubChem introduced a dynamic web traffic throttling approach to enforce the usage policies (https://pubchemdocs.ncbi.nlm.nih.gov/programmatic-access$_RequestVolumeLimitations). During periods of excessive demand, this dynamic throttling algorithm tightens the usage policies to maintain accessibility to all users. Users should moderate the speed at which requests are sent to PubChem, according to the traffic status and usage limits, which can be found in specialized HTTP response headers for web requests. Requests exceeding the usage limits are rejected with an HTTP 503 error. Those who continuously exceed the limits will be temporarily blocked for a period of time. More detailed information on the usage polices and dynamic traffic throttling is provided in our recent publication (28).
SUMMARY
Over the past few years, a substantial amount of new data has been added to PubChem. Thanks to integration of data from SpectraBase and MoNA, PubChem now contains a wide range of spectral information for about 600 000 compounds. Data contribution from Springer Nature has enabled PubChem to provide 26 million links between 600 000 chemicals and four million papers. In addition, the scope of PubChem annotation data has been expanded to food and agricultural chemicals.
PubChem has also released several new services. The PubChem Target View page for a gene or protein target provides a target-centric view of all information available in PubChem for the target as well as annotations collected from many authoritative sources. The Bioactivity dyad page presents bioactivity data of a chemical tested in a particular assay or against a particular target. The Patent View summarizes chemicals mentioned in a given patent, along with other information, such as patent title, abstract, application/publication dates, applicant, inventor, and patent classification. PubChem Sources page help users understand where data in PubChem come from. PubChem Widgets 2.0f allows one to display PubChem data on their own webpages. PUG-View enable users to programmatically access annotations presented on the Summary or Record page of PubChem records. PubChem introduced a dynamic traffic throttling system to help maximize uptime and request handling speed.
With researchers in mind, PubChem continues to improve and develop tools and services that enable rapid access to information. This includes new efforts to enhance search and reorganize content with a focus towards summarizing what is known along with the evidence for assertions about the associations between biologically important entities (such as chemical-gene and chemical-disease associations). As a part of PubChem's continuing modernization efforts, some older PubChem tools may be deprecated or substantially modified to keep up with changing technology and the demands of the modern researcher.
DATA AVAILABILITY
All PubChem data, tools, and services are provided to the public free of charge.
ACKNOWLEDGEMENTS
We appreciate the hundreds of data contributors for making their data openly accessible within PubChem. Special thanks go to the entire NCBI staff (especially to the help desk and systems support teams).
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al. . PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Bryant S.H., Cheng T., Wang J., Gindulyte A., Shoemaker B.A., Thiessen P.A., He S., Zhang J.. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017; 45:D955–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S. Getting the most out of PubChem for virtual screening. Expert Opin. Drug Discov. 2016; 11:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hähnke V.D., Kim S., Bolton E.E.. PubChem chemical structure standardization. J. Cheminform. 2018; 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S., Thiessen P.A., Cheng T., Yu B., Shoemaker B.A., Wang J.Y., Bolton E.E., Wang Y.L., Bryant S.H.. Literature information in PubChem: associations between PubChem records and scientific articles. J. Cheminform. 2016; 8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meija J., Coplen T.B., Berglund M., Brand W.A., De Bievre P., Groning M., Holden N.E., Irrgeher J., Loss R.D., Walczyk T. et al. . Atomic weights of the elements 2013 (IUPAC Technical Report). Pure Appl. Chem. 2016; 88:265–291. [Google Scholar]
- 7.Meija J., Coplen T.B., Berglund M., Brand W.A., De Bievre P., Groning M., Holden N.E., Irrgeher J., Loss R.D., Walczyk T. et al. . Isotopic compositions of the elements 2013 (IUPAC Technical Report). Pure Appl. Chem. 2016; 88:293–306. [Google Scholar]
- 8.Audi G., Kondev F.G., Wang M., Huang W.J., Naimi S.. The NUBASE2016 evaluation of nuclear properties. Chin. Phys. C. 2017; 41:030001. [Google Scholar]
- 9.Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrian-Uhalte E. et al. . The ChEMBL database in 2017. Nucleic Acids Res. 2017; 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al. . DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., Gray A.J.G., Bruce L., Alexander S.P.H., Anderton S. et al. . The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018; 46:D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R. et al. . Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015; 43:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. . Gene Ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbon S., Dietze H., Lewis S.E., Mungall C.J., Munoz-Torres M.C., Basu S., Chisholm R.L., Dodson R.J., Fey P., Thomas P.D. et al. . Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S., Bruford E.A.. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017; 45:D619–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R. et al. . UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose P.W., Prlic A., Altunkaya A., Bi C.X., Bradley A.R., Christie C.H., Di Costanzo L., Duarte J.M., Dutta S., Feng Z.K. et al. . The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A., Bo Y., Han L.Y., He J.E., Lanczycki C.J., Lu S.N., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. et al. . CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halavi M., Maglott D., Gorelenkov V., Rubinstein W.. MedGen. The NCBI Handbook [Internet]. 2013; 2nd edBethesda: National Center for Biotechnology Information (US). [Google Scholar]
- 21.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geer L.Y., Marchler-Bauer A., Geer R.C., Han L.Y., He J., He S.Q., Liu C.L., Shi W.Y., Bryant S.H.. The NCBI BioSystems database. Nucleic Acids Res. 2010; 38:D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madej T., Lanczycki C.J., Zhang D.C., Thiessen P.A., Geer R.C., Marchler-Bauer A., Bryant S.H.. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014; 42:D297–D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papadatos G., Davies M., Dedman N., Chambers J., Gaulton A., Siddle J., Koks R., Irvine S.A., Pettersson J., Goncharoff N. et al. . SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 2016; 44:D1220–D1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heifets A., Jurisica I.. SCRIPDB: a portal for easy access to syntheses, chemicals and reactions in patents. Nucleic Acids Res. 2012; 40:D428–D433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilson M.K., Liu T.Q., Baitaluk M., Nicola G., Hwang L., Chong J.. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016; 44:D1045–D1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Thiessen P.A., Bolton E.E., Bryant S.H.. PUG-SOAP and PUG-REST: web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015; 43:W605–W611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., Thiessen P.A., Cheng T.J., Yu B., Bolton E.E.. An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Res. 2018; 46:W563–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All PubChem data, tools, and services are provided to the public free of charge.