The Alliance of Genome Resources: Building a Modern Data Ecosystem for Model Organism Databases (original) (raw)
Abstract
Model organisms are essential experimental platforms for discovering gene functions, defining protein and genetic networks, uncovering functional consequences of human genome variation, and for modeling human disease. For decades, researchers who use model organisms have relied on Model Organism Databases (MODs) and the Gene Ontology Consortium (GOC) for expertly curated annotations, and for access to integrated genomic and biological information obtained from the scientific literature and public data archives. Through the development and enforcement of data and semantic standards, these genome resources provide rapid access to the collected knowledge of model organisms in human readable and computation-ready formats that would otherwise require countless hours for individual researchers to assemble on their own. Since their inception, the MODs for the predominant biomedical model organisms [Mus sp. (laboratory mouse), Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Danio rerio, and _Rattus norvegicus_] along with the GOC have operated as a network of independent, highly collaborative genome resources. In 2016, these six MODs and the GOC joined forces as the Alliance of Genome Resources (the Alliance). By implementing shared programmatic access methods and data-specific web pages with a unified “look and feel,” the Alliance is tackling barriers that have limited the ability of researchers to easily compare common data types and annotations across model organisms. To adapt to the rapidly changing landscape for evaluating and funding core data resources, the Alliance is building a modern, extensible, and operationally efficient “knowledge commons” for model organisms using shared, modular infrastructure.
Keywords: model organism databases, bioinformatics, data stewardship, database sustainability
A Brief History of Model Organism Databases and the Gene Ontology Consortium
BECAUSE many basic biological processes and molecular mechanisms are shared across all extant organisms, discoveries in diverse nonprimate organisms can reveal fundamental properties of the homologous biological processes in humans. Model organisms, including Mus sp. (laboratory mouse), Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Danio rerio, and Rattus norvegicus, and model systems less commonly used have provided insights into the biological processes that underlie human health and disease, and have contributed to the development of diagnoses and treatments for genetic diseases (Iannaccone and Jacob 2009; Phillips and Westerfield 2014; Hamza et al. 2015; Kachroo et al. 2015; Strange 2016; Ugur et al. 2016; Bonini and Berger 2017; Golden 2017; Sen and Cox 2017; Wangler et al. 2017; Apfeld and Alper 2018; Ingham 2018; Nadeau and Auwerx 2019; Smith et al. 2019).
Model organism databases (MODs) have played a central role in the success of animal models in basic and biomedical research for decades by providing ready access to knowledge about genome features, their functions, and their associated phenotypes. MODs obtain and continually update this information through expert curation and integration of heterogeneous data and information from peer-reviewed scientific literature, and from direct data submissions. To assist researchers in finding appropriate models for studying biological mechanisms that contribute to complex phenotypes and disease, MODs provide access to inventories of biological reagents that are available from stock centers and strain repositories. They also maintain linkages to relevant data available in scores of other genome-centric bioinformatics resources and sequence archives, such as UniProtKB (UniProt Consortium 2019) and GenBank (Benson et al. 2018). The MODs work closely with their respective organism-specific research communities to define nomenclature and data format standards, and they serve as the authoritative sources of most organism-specific gene, phenotype, and disease annotations (Table 1). Acknowledgments of the MODs and the Gene Ontology Consortium (GOC) in the peer-reviewed scientific literature demonstrate that these resources are widely used to support science funded across all National Institutes of Health (NIH) Institutes and have global impact. These resources also have been leveraged heavily by bioinformatics initiatives using comparative biology approaches for functional genomics, including MARRVEL (Model organism Aggregated Resources for Rare Variant ExpLoration) (Wang et al. 2017), the Monarch Initiative (Mungall et al. 2017), GeneWeaver (Bubier et al. 2017), Gene2Function (Hu et al. 2017), and modEnrichr (Kuleshov et al. 2019). As noted by Oliver et al. (2016), “Without the systematic organization of the MODs, each of our research efforts would be drastically impeded and, in some cases, impossible, slowing the pace of discovery and reducing the efficient use of NIH funding.”
Table 1. The founding members of the Alliance of Genome Resources and the data for which the resource is the authoritative source: the NHGRI at the NIH is the primary funder for all of the resources except for Rat Genome Database, where the primary funding comes from the National Heart Lung Blood Institute.
| Genome resource | Year founded | Authoritative data/annotations |
|---|---|---|
| Mouse Genome Database (MGD): http://www.informatics.jax.org/; Bult et al. (2019) | 1989 | Mouse gene, allele, and strain nomenclature; gene function (GO) annotations; phenotype annotations; mouse models of human disease; unified genome feature catalog for the mouse reference genome |
| FlyBase: https://flybase.org/; Thurmond et al. (2019) | 1992 | Drosophila gene and allele nomenclature; gene function annotations (GO); protein annotation; phenotype annotations; fly models of human disease |
| Saccharomyces Genome Database (SGD): https://yeastgenome.org/; Cherry et al. (2012) | 1993 | S. cerevisiae reference genome sequence; reference proteome and chromosomal feature annotations; standardized nomenclature for gene names; gene product annotations; gene function (GO) annotations; phenotype and human disease associations; regulatory networks; gene expression patterns; Metabolic pathways; curation of all S. cerevisiae published literature. |
| Zebrafish Information Network (ZFIN): https://zfin.org/; Westerfield et al. (1997) | 1994 | Zebrafish gene, allele, and strain nomenclature; gene function (GO) annotations; gene expression annotations; phenotype annotations; zebrafish models of human disease; unified genome feature catalog for the zebrafish reference genome; reagents; catalog of Zebrafish researchers |
| Gene Ontology Consortium (GOC): http://geneontology.org/: The Gene Ontology (2019) | 1998 | GO (classification of gene functions) terms and relationships among terms for Biological Process, Molecular Function, and Cellular Component; GO annotations from multiple sources |
| Rat Genome Database (RGD): https://rgd.mcw.edu/; Laulederkind et al. (2018) | 1999 | Rat gene, allele, QTL, cell line, and strain nomenclature; gene function (GO) annotations; human disease, phenotype, and pathway annotations; quantitative phenotype measurement records, including expected ranges for individual rat strains. |
| WormBase: https://www.wormbase.org/; Lee et al. (2018) | 2000 | C. elegans reference genome sequence and curated gene structures; nomenclature for numerous data types, including genes and alleles; gene function (GO) annotation; expression and interaction annotation; ontologies for nematode phenotypes and development; catalog of C. elegans researchers. |
Common needs of the different MOD user communities have led to collaborations among the MODs to develop novel and important centralized genome resources. Gene Ontology (GO), for example, was launched to annotate gene product function, biological processes, and cellular location across different organisms with common, well-defined terms (Ashburner et al. 2000). Start-up funding from AstraZeneca, along with stable funding provided subsequently by the National Human Genome Research Institute (NHGRI), supported the centralized development of the GO and related software tools, as well as coordinated gene function curation efforts among the first GOC members: FlyBase, the Mouse Genome Database (MGD), and the Saccharomyces Genome Database. The GOC has since grown to include > 30 active members (see http://geneontology.org/docs/annotation-contributors/) and is one of the most cited resources in biomedicine (Duck et al. 2016). It is a crucial resource for the interpretation of high-throughput experimental data, and cross-species data retrieval and aggregation (Blake and Bult 2006). The GO has also spurred the development of a number of other ontologies for related biological domains, including Cell Ontology (Diehl et al. 2016) and Uberon, the multi-species anatomy ontology (Mungall et al. 2012).
The fundamental data management principles upon which MODs and the GOC were built were designed to promote “rigor and reproducibility” in biomedical research, through the generation and maintenance of stable references to biological entities and annotations. In recent times, these concepts are better known as FAIR principles (Findability, Accessibility, Interoperability, and Reusability) (Wilkinson et al. 2016). These principles remain a constant at the core of operations for MODs and the GOC, even as the resources continually adapt to accommodate new data types, curation methods, and data management technologies.
The Changing Landscape for Sustaining Core Data Resources
Although several of the current major MODs existed prior to the genome era (Table 1), a large investment was made in genome knowledgebases by the NIH and, in particular, the NHGRI, starting around the time of the Human Genome Project in the early 1990s. These investments were made in recognition of the importance of model organisms for understanding the biology of the human genome and for advancing the application of genomics to medical practice. An NIH-sponsored workshop focusing specifically on the importance of nonmammalian model organisms was held in February 1999 (see https://web.archive.org/web/20000818110738/https://www.nih.gov/science/models/nmm/) following a similar workshop organized by the National Cancer Institute in 1997 (see https://web.archive.org/web/20000818162500/http://www.nih.gov/science/models/nmm/nci_nmm_report.html). The executive summary from the 1999 meeting emphasized the critical need for genome sequencing, molecular and organismal reagents, and public databases for nonmammalian model organisms to support the interpretation of the human genome.
Early in 2016, the NHGRI, the primary funder of most MODs and the GOC, announced their intent to scale back funding for these community genome resources by 30% by Fiscal Year 2021. The leaders of the MODs and the GOC were urged by the NHGRI to restructure the organization, management, and operations of their resources to achieve substantial cost savings (Hayden 2016; Kaiser 2016). The main justifications for the mandated changes were threefold. First, there was a concern that the lack of uniformity in user interfaces across the different resources had resulted in unintentional “siloing” of information because users had to navigate different search and display options for common data types at the different MOD websites. For computational biologists, the lack of unified programmatic data access methods meant that unique code had to be written for each database to retrieve similar types of data and annotations. Second, there was a perception that there were unnecessary redundancies in operations and infrastructure, due to the independent and distributed nature of the genome resources. Centralization of infrastructure was seen as a means to reduce the overall operational and management costs of the resources. Finally, while recognizing the critical importance of these resources, the NHGRI argued that the ongoing financial commitment to these resources was restricting the investments that they could make in new areas of genome research.
In May of 2016, the principal investigators from six MODs and the GOC presented a concept for a unified MOD/GOC initiative to NIH program officials and their external scientific advisors. Following this meeting, the MOD/GOC coalition submitted a formal proposal to fund the initial steps needed to implement the proposed framework. This proposal was awarded as an administrative supplement to the WormBase grant in September of 2016, formally launching the Alliance.
The response of the research community to the NHGRI’s announcement of reduced funding for the MODs/GOC was one of concern and alarm. Under the auspices of the Genetics Society of America, the Society for Developmental Biology, and the American Society of Cell Biology, a Statement of Support for the MODs was published that urged the NHGRI/NIH to reconsider the funding cutbacks. The Statement highlighted the importance of the MODs in supporting basic research and discovery, and advocated for continued “adequate and sustained funding” for the resources. The statement was signed by over 11,000 scientists (see Poston 2016; http://genestogenomes.org/action-alert-support-model-organism-database-funding/), including 12 Nobel Laureates and 57 members of the National Academy of Sciences. It was presented to the NIH Director, Francis Collins, at The Allied Genetics Conference (TAGC) in the summer of 2016 (Organizers of The Allied Genetics Conference 2016).
As a follow-up to the TAGC meeting, NIH program officials and external advisors, community stakeholders, and representatives of the MODs and the GOC assembled for a meeting on genome resource sustainability in Bethesda in March 2017. At this meeting, the plans and progress of the Alliance were reported and discussed. Although no specific plans were presented at this meeting for a new evaluation and funding model for community resources, NHGRI Director Eric Green reported on early stage national and international discussions focused on developing strategies for sustainable funding of core data resources. Patricia Brennan, the newly appointed National Library of Medicine (NLM) Director, acknowledged the importance of MODs, and affirmed the NLM’s commitment to data standards and interoperability.
The NIH released a strategic plan for data science in June 2018 (see https://www.nih.gov/news-events/news-releases/nih-releases-strategic-plan-data-science). The plan outlines the need and vision for “modernizing the NIH-funded biomedical data science ecosystem,” addresses the challenges of defining meaningful criteria with which to evaluate core community resources, and acknowledges the need for evaluation criteria specifically for bioinformatics resources. Although the plan touches on many of the important challenges for data science in biomedical research, a corresponding tactical plan for sustainable funding of core resources has yet to emerge.
The Alliance of Genome Resources
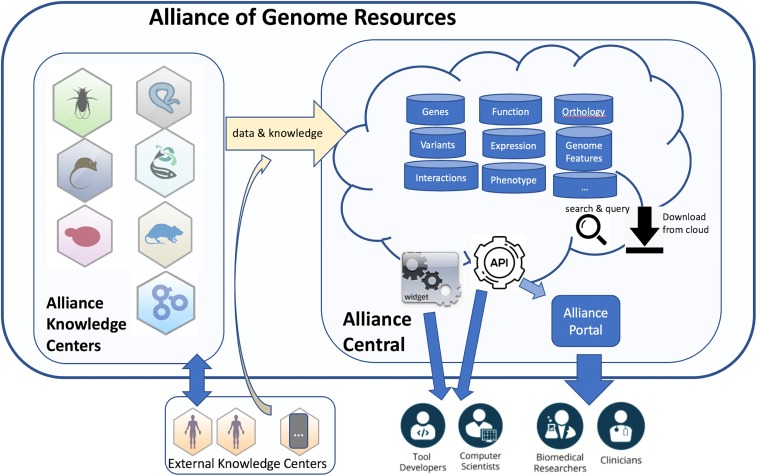
The Alliance of Genome Resources is more than a formal consortium among the MODs and the GOC. It represents a significant departure from a mostly decentralized approach to knowledgebase development and maintenance to a highly centralized and coordinated effort. Organizationally, the Alliance has two interdependent functional units: Alliance Central and Alliance Knowledge Centers (Figure 1). Alliance Central is responsible for developing and maintaining the software platform and shared modular infrastructure, and for the coordination of data harmonization activities across the Knowledge Centers. The coordination of infrastructure development reduces redundancy in systems administration, software development, and ensures a unified “look and feel” for access and display of data types in common across diverse model organisms. Alliance Knowledge Centers are responsible for expert curation of data and for submission of data to Alliance Central using common standardized data formats. Knowledge Centers also are responsible for organism-specific user support activities and for providing access to data types not yet supported by Alliance Central.
Figure 1.
The Alliance of Genome Resources is organized into Knowledge Centers (expert curation, development of ontologies and standards, and data integration) and Alliance Central (data management and delivery, software tools, and widgets). Alliance Central provides centralized infrastructure support for Knowledge Centers. Knowledge Centers are federated to support maximally effective organism-specific data acquisition and curation. Shared standards for knowledge representation and data formats allow for unification of Alliance Knowledge Centers with external knowledge bases that are relevant to the Alliance mission but are not formal Alliance members. API, application programming interface.
The Alliance of Genome Resources serves the same diverse research communities supported by the existing collective of model organism genome resources including: (i) human geneticists and clinical researchers who want access to all model organism data, which are the main sources of experimental annotation of human genes through orthology; (ii) basic scientists who use specific model organisms to investigate fundamental biology; (iii) computational biologists and data scientists who need access to standardized, well-structured data, both big and small; and (iv) educators and students. As a consortium, the Alliance is a powerful advocate for model organism research and will serve these diverse user communities even better than before. Model organism researchers will benefit from streamlined development and coordinated delivery of access to new data types and user interfaces. Model organisms with smaller user communities will be able to leverage Alliance infrastructure to enhance their impact in advancing genome biology and translational research. Computational biologists and data scientists will benefit from the centralized data access and common Application Programming Interfaces.
Since its official launch in 2016, the Alliance has made substantial progress toward unified access to common data types across different organisms and the development of a scalable data ecosystem for model organism knowledgebases (Table 2). Examples of the accomplishments of the Alliance to date include: (i) a single integrated Alliance orthology gene set for comparative genomics of humans and model organisms, based on the work of the Quest for Orthologs Consortium (Glover et al. 2019); (ii) adoption of the Disease Ontology as the common annotation standard for annotating human disease association; (iii) a ribbon visualization widget to display summary annotations for gene function, phenotype, and expression developed initially by the MGD (Bult et al. 2016) that has been implemented by Alliance developers as a reusable web component for displaying annotations across multiple organisms, and (iv) a computational method developed by WormBase for automatically generating brief, readable summaries of gene function from ontology annotations, which is now used across the Alliance members to generate gene summaries for model organisms and human. A recent publication on the functionality currently supported by the Alliance website illustrates how researchers can search the resource by gene symbols, gene function terms, and disease terms, and then review annotations from all six model organisms and human using interfaces that share a common look and feel (Alliance of Genome Resources Consortium 2019).
Table 2. Examples of the accomplishments of the Alliance of Genome Resources to date in the areas of organization, process, data, and interfaces and how these accomplishments benefit the research community.
| Accomplishment | Community benefit |
|---|---|
| Organization: Common project management and governance structure | Ability to leverage unique capabilities and expertise to enhance genome resources |
| Organization: Centralized user Help Desk | Single point of access for inquiries about data for any organism in the Alliance |
| Organization: Coordinated software development | Rapid propagation of access to new data types and interfaces across model organisms |
| Process: Data harmonization | Essential for developing user interfaces with a unified “look and feel” for common data types |
| Process: Automated processes for concise, human-readable summaries of gene function | A short, human readable summary of gene function standardized across all model organisms in the Alliance |
| Data: Common set of orthologs | Supports comparisons of gene function, phenotype, and disease annotations among model organisms and with human data |
| Data: Common protein–protein interaction data | Leverage existing community resources to provide a common set of PPI data for all model organisms in the Alliance (Orchard et al. 2012; Oughtred et al. 2019) |
| Interface: Sequence display widget | Common graphical representation of transcripts for a gene |
| Interface: JBrowse genome browser | Adoption of externally developed software as the standard genome browser for all model organisms Skinner et al. (2009) |
| Interface: “Ribbon” widget for visualizing gene function and expression annotation summaries | Unified visualization paradigm for annotation summary information across all model organisms in the Alliance |
| Interface: Common web pages for genes and diseases | Consistent organization of common data types across all model organisms in the Alliance |
| Interface: Common application programming interface for common data types | Single point of programmatic access for common data types across all model organisms in the Alliance |
Future Directions for the Alliance and Core Data Resource Sustainability
The transformational potential of the Alliance of Genome Resources is already being realized in operational efficiencies and enhanced user experiences, driven by an enhanced capacity for rapid delivery of new data types and user interfaces designed to facilitate comparative biology. The approach to infrastructure development within the Alliance reflects the central principles articulated in the NIH’s data science strategic plan as well as the requirements for core community resources outlined by the European life-sciences Infrastructure for biological information (ELIXIR) program initiative (Durinx et al. 2016). The Alliance builds on previous successful cross-MOD projects and related initiatives, including the Generic Model Organism Database project (Stein et al. 2002; O’Connor et al. 2008) and InterMine (Lyne et al. 2015). Tools and interfaces developed by the Alliance are architected for reuse by others. The Alliance-developed “Sequence Feature Viewer” widget, for example, has been adopted by the Monarch Initiative (Mungall et al. 2017) for use at their website. Further, the Alliance will seek to adopt, rather than develop, tools and interfaces. For example, the Alliance is using JBrowse (Skinner et al. 2009) as a common genome browser application and are working with the JBrowse development team to add new functionality.
Eventually, the Alliance resource will reflect the union of data and functionality currently supported by individual MODs and the GOC, but this will take several years to achieve because it is critical that this goal be accomplished without sacrificing existing quality of service and timeliness of data updates to organism-specific user communities. For the near term, the Alliance web portal (www.alliancegenome.org) and the original, pre-Alliance MOD and GOC websites and infrastructure will coexist. Gradually, interfaces and resources developed by the Alliance are being deployed by the individual MODs. As new shared components are developed within Alliance Central, each MOD will retire its existing infrastructure and adopt the shared components. By 2024, we envision that the concept of “develop once, use by all” will be the standard operating procedure for data types and software tools shared among all Alliance Knowledge Centers.
The vision and roadmap for the Alliance are clear, and the initiative will be funded by the NHGRI for at least the next 5 years (2019–2024). However, there is significant uncertainty regarding long-term funding for the Alliance and all core community data resources. Ideas for funding models to reduce reliance on federal grants include public funding, third party payers, and commercialization (Anderson and Global Life Science Data Resources Working Group 2017; Gabella et al. 2017). The international nature of community resources and their user communities will likely require a mixed model to address the questions of what constitutes a core data resource, and how to sustain it.
Just as there are well-accepted principles for data management (e.g., FAIR) to support data reuse, decisions regarding funding for core community data resources should be guided by principles of data stewardship that extend beyond initial funding for data generation and short-term support for project-specific data coordination centers. For data and information that are of broad utility to the research community, data stewardship practices by the agencies that fund data generation should reflect a commitment to Sustained data Access For Everyone (SAFE) and be measured by adherence to—and long-term financial support of—essential data stewardship practices (Peng et al. 2015), including:
- sustainable and open data access (both programmatic and web-based),
- preservation of data and annotation quality,
- maintenance of data integrity and usability,
- traceability of provenance for data and annotations,
- clear guidelines for permissible data use, and
- support for community outreach and training.
The MODs, GOC, and the Alliance are data stewards for the global research community. Our efforts ensure that best practices for data management and data stewardship principles are enforced. In turn, this work preserves, and enhances, the impact of the significant financial investment made by government agencies and foundations in biological and biomedical research initiatives.
Acknowledgments
We thank all members of the Alliance of Genome Resources, the staff of the MODs and GOC, and the Alliance Scientific Advisory Board for their contributions to developing the vision and approach for the Alliance. V.D.F. and R.F. contributed to this manuscript in their official roles as program coordinators for the National Institutes of Health National Human Genome Research Institute.
The Alliance of Genome Resources Consortium
Carol J. Bult
The Jackson Laboratory for Mammalian Genetics,
Bar Harbor, Maine 04609
(Orcid ID: 0000-0001-9433-210X)
Judith A. Blake
The Jackson Laboratory for Mammalian Genetics,
Bar Harbor, Maine 04609
(Orcid ID: 0000-0001-8522-334X)
Brian R. Calvi
Department of Biology, Indiana University,
Bloomington, Indiana 47405
(Orcid ID: 0000-0001-5304-0047)
J. Michael Cherry
Department of Genetics, Stanford University,
Palo Alto, California 94305
(Orcid ID: 0000-0001-9163-5180)
Valentina DiFrancesco
National Human Genome Research Institute,
Bethesda, Maryland 20892
Robert Fullem
National Human Genome Research Institute,
Bethesda, Maryland 20892
(Orcid ID: 0000-0003-0141-7767)
Kevin L. Howe
European Bioinformatics Institute (EMBL-EBI), European Molecular Biology Laboratory,
Hinxton, Cambridgeshire CB10 1SD, UK
(Orcid ID: 0000-0002-1751-9226)
Thom Kaufman
Indiana University
Bloomington, Indiana 47405
(Orcid ID: 0000-0003-1406-7671)
Chris Mungall
Lawrence Berkeley National Laboratory,
Berkeley, California 94720
(Orcid ID: 0000-0002-6601-216)
Norbert Perrimon
Department of Genetics, Harvard University,
Boston, Massachusetts 02138
(Orcid ID: 0000-0001-7542-472X)
Mary Shimoyama
Medical College of Wisconsin, Madison, Wisconsin 53226
(Orcid ID: 0000-0003-1176-0796)
Paul W. Sternberg
Division of Biology and Biological Engineering,
California Institute of Technology,
Pasadena, California 91125
(Orcid ID: 0000-0002-7699-0173)
Paul Thomas
Keck School of Medicine, University of Southern California,
Los Angeles, California 90089
(Orcid ID: 0000-0002-9074-3507)
Monte Westerfield
Department of Biology, University of Oregon, Eugene,
Oregon 97403
Footnotes
Communicating editor: M. Johnston
2
A full list of members is provided at the end of this article.
Literature Cited
- Anderson W. P.; Global Life Science Data Resources Working Group , 2017. Data management: a global coalition to sustain core data. Nature 543: 179 10.1038/543179a [DOI] [PubMed] [Google Scholar]
- Apfeld J., and Alper S., 2018. What can we learn about human disease from the nematode C. elegans? Methods Mol. Biol. 1706: 53–75. 10.1007/978-1-4939-7471-9_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J. et al. , 2018. GenBank. Nucleic Acids Res. 46: D41–D47. 10.1093/nar/gkx1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J. A., and Bult C. J., 2006. Beyond the data deluge: data integration and bio-ontologies. J. Biomed. Inform. 39: 314–320. 10.1016/j.jbi.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Bonini N. M., and Berger S. L., 2017. The sustained impact of model organisms-in genetics and epigenetics. Genetics 205: 1–4. 10.1534/genetics.116.187864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier J. A., Langston M. A., Baker E. J., and Chesler E. J., 2017. Integrative functional genomics for systems genetics in GeneWeaver.org. Methods Mol. Biol. 1488: 131–152. 10.1007/978-1-4939-6427-7_6 [DOI] [PubMed] [Google Scholar]
- Bult C. J., Eppig J. T., Blake J. A., Kadin J. A., Richardson J. E. et al. , 2016. Mouse genome database 2016. Nucleic Acids Res. 44: D840–D847. 10.1093/nar/gkv1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C. J., Blake J. A., Smith C. L., Kadin J. A., Richardson J. E. et al. , 2019. Mouse genome database (MGD) 2019. Nucleic Acids Res. 47: D801–D806. 10.1093/nar/gky1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M., Hong E. L., Amundsen C., Balakrishnan R., Binkley G. et al. , 2012. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40: D700–D705. 10.1093/nar/gkr1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl A. D., Meehan T. F., Bradford Y. M., Brush M. H., Dahdul W. M. et al. , 2016. The cell ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics 7: 44 10.1186/s13326-016-0088-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duck G., Nenadic G., Filannino M., Brass A., Robertson D. L. et al. , 2016. A survey of bioinformatics database and software usage through mining the literature. PLoS One 11: e0157989 10.1371/journal.pone.0157989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinx C., McEntyre J., Appel R., Apweiler R., Barlow M. et al. , 2016. Identifying ELIXIR core data resources. F1000Res. 5: ELIXIR-2422. 10.12688/f1000research.9656.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabella C., Durinx C., and Appel R., 2017. Funding knowledgebases: towards a sustainable funding model for the UniProt use case. F1000Res. 6: ELIXIR-2051. 10.12688/f1000research.12989.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover N., Dessimoz C., Ebersberger I., Forslund S. K., Gabaldon T. et al. , 2019. Advances and applications in the quest for Orthologs. Mol. Biol. Evol. 36: 2157–2164. 10.1093/molbev/msz150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A., 2017. From phenologs to silent suppressors: identifying potential therapeutic targets for human disease. Mol. Reprod. Dev. 84: 1118–1132. 10.1002/mrd.22880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza A., Tammpere E., Kofoed M., Keong C., Chiang J. et al. , 2015. Complementation of yeast genes with human genes as an experimental platform for functional testing of human genetic variants. Genetics 201: 1263–1274. 10.1534/genetics.115.181099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden E. C., 2016. Concern over funding cuts for model organism databases. Nature. DOI: 10.1038/nature.2016.20134. [Google Scholar]
- Hu Y., Comjean A., Mohr S. E.; FlyBase Consortium, and Perrimon N., 2017. Gene2Function: an integrated online resource for gene function discovery. G3 (Bethesda) 7: 2855–2858. 10.1534/g3.117.043885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannaccone P. M., and Jacob H. J., 2009. Rats! Dis. Model. Mech. 2: 206–210. 10.1242/dmm.002733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., 2018. From Drosophila segmentation to human cancer therapy. Development 145: dev168898. 10.1242/dev.168898 [DOI] [PubMed] [Google Scholar]
- Kachroo A. H., Laurent J. M., Yellman C. M., Meyer A. G., Wilke C. O. et al. , 2015. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348: 921–925. 10.1126/science.aaa0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J., 2016. BIOMEDICAL RESOURCES. Funding for key data resources in jeopardy. Science 351: 14 10.1126/science.351.6268.14 [DOI] [PubMed] [Google Scholar]
- Kuleshov M. V., Diaz J. E. L., Flamholz Z. N., Keenan A. B., Lachmann A. et al. , 2019. modEnrichr: a suite of gene set enrichment analysis tools for model organisms. Nucleic Acids Res. 47: W183–W190. 10.1093/nar/gkz347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulederkind S. J. F., Hayman G. T., Wang S. J., Smith J. R., Petri V. et al. , 2018. A primer for the rat genome database (RGD). Methods Mol. Biol. 1757: 163–209. 10.1007/978-1-4939-7737-6_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y. N., Howe K. L., Harris T. W., Arnaboldi V., Cain S. et al. , 2018. WormBase 2017: molting into a new stage. Nucleic Acids Res. 46: D869–D874. 10.1093/nar/gkx998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R., Sullivan J., Butano D., Contrino S., Heimbach J. et al. , 2015. Cross-organism analysis using InterMine. Genesis 53: 547–560. 10.1002/dvg.22869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C. J., Torniai C., Gkoutos G. V., Lewis S. E., and Haendel M. A., 2012. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 13: R5 10.1186/gb-2012-13-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C. J., McMurry J. A., Kohler S., Balhoff J. P., Borromeo C. et al. , 2017. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 45: D712–D722. 10.1093/nar/gkw1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., and Auwerx J., 2019. The virtuous cycle of human genetics and mouse models in drug discovery. Nat. Rev. Drug Discov. 18: 255–272. 10.1038/s41573-018-0009-9 [DOI] [PubMed] [Google Scholar]
- O’Connor B. D., Day A., Cain S., Arnaiz O., Sperling L. et al. , 2008. GMODWeb: a web framework for the generic model organism database. Genome Biol. 9: R102 10.1186/gb-2008-9-6-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., Lock A., Harris M. A., Nurse P., and Wood V., 2016. Model organism databases: essential resources that need the support of both funders and users. BMC Biol. 14: 49 10.1186/s12915-016-0276-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J. et al. , 2012. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods 9: 345–350 (erratum: Nat. Methods 9: 626). 10.1038/nmeth.1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organizers of The Allied Genetics Conference 2016 Meeting Report: The Allied Genetics Conference 2016. G3 (Bethesda) 6: 3765–3786. . doi: 10.1534/g3.116.036848. [DOI] [Google Scholar]
- Oughtred R., Stark C., Breitkreutz B. J., Rust J., Boucher L. et al. , 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47: D529–D541. 10.1093/nar/gky1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Privette J. L., Kearns E. J., Ritchey N. A., and Ansari S., 2015. A unified framework for measuring stewardship practices applied to digital environmental datasets. Data Sci. J. 13: 231–253. 10.2481/dsj.14-049 [DOI] [Google Scholar]
- Phillips J. B., and Westerfield M., 2014. Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis. Model. Mech. 7: 739–743. 10.1242/dmm.015545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston C., 2016. Action Alert: Support model organism database funding. Genes to Genomes: A Blog from the Genetics society of America. Available at: http://genestogenomes.org/action-alert-support-model-organism-database-funding. Accessed: October 11, 2019. PMCID: PMC5144950.
- Sen A., and Cox R. T., 2017. Fly models of human diseases: Drosophila as a model for understanding human mitochondrial mutations and disease. Curr. Top. Dev. Biol. 121: 1–27. 10.1016/bs.ctdb.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Skinner M. E., Uzilov A. V., Stein L. D., Mungall C. J., and Holmes I. H., 2009. JBrowse: a next-generation genome browser. Genome Res. 19: 1630–1638. 10.1101/gr.094607.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. R., Bolton E. R., and Dwinell M. R., 2019. The rat: a model used in biomedical research, pp. 1–41 in Rat Genomics. Methods in Molecular Biology, edited by Hayman G. T., Smith J. R., Dwinell M. R., and Shimoyama M.. Springer-Verlag, New York: 10.1007/978-1-4939-9581-3_1 [DOI] [PubMed] [Google Scholar]
- Stein L. D., Mungall C., Shu S., Caudy M., Mangone M. et al. , 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12: 1599–1610. 10.1101/gr.403602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange K., 2016. Drug discovery in fish, flies, and worms. ILAR J. 57: 133–143. 10.1093/ilar/ilw034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliance of Genome Resources Consortium , 2019. Alliance of Genome Resources Portal: unified model organism research platform. Nucleic Acids Res. DOI: 10.1093/nar/gkz813. 10.1093/nar/gkz813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2019. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47: D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S. et al. , 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur B., Chen K., and Bellen H. J., 2016. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 9: 235–244. 10.1242/dmm.023762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium , 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47: D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Al-Ouran R., Hu Y., Kim S. Y., Wan Y. W. et al. , 2017. MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 100: 843–853. 10.1016/j.ajhg.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler M. F., Yamamoto S., Chao H. T., Posey J. E., Westerfield M. et al. , 2017. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207: 9–27. 10.1534/genetics.117.203067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M., Doerry E., Kirkpatrick A. E., Driever W., and Douglas S. A., 1997. An on-line database for zebrafish development and genetics research. Semin. Cell Dev. Biol. 8: 477–488. 10.1006/scdb.1997.0173 [DOI] [PubMed] [Google Scholar]
- Wilkinson M. D., Dumontier M., Aalbersberg I. J., Appleton G., Axton M. et al. , 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3: 160018 [corrigenda: Sci. Data 6: 6 (2019)]. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]