AT-AC Pre-mRNA Splicing Mechanisms and Conservation of Minor Introns in Voltage-Gated Ion Channel Genes (original) (raw)
Shortly after the discovery of split genes in 1977, a conserved sequence feature at both ends of cellular and viral introns was recognized, i.e., the presence of GT at the 5′ splice site and AG at the 3′ splice site, giving rise to the so-called GT-AG rule (8). This rule holds in most cases, but exceptions have been found. For example, GC is occasionally found at the 5′ end of certain introns (Table 1) (38). GC-AG introns are processed by the same splicing pathway as conventional GT-AG introns (3). It had long been assumed that removal of all introns from eukaryotic pre-mRNAs took place by the same splicing pathway, until recent developments demonstrated the existence of a second pre-mRNA splicing pathway.
TABLE 1.
Compilation of GC-AG intronsa
| Gene product | Intron no. | 5′ splice site | Size (nt) |
|---|---|---|---|
| SCA2c | 9 | AGgcaagt | 6,600 |
| Protein Zd | 2 | CTgcaagt | NAb |
| AGLe | 17 | AGgcaaga | 400 |
| LAMA4f | 7 | AGgcatgg | NA |
| s-Lamining | 1 | AGgcagag | 86 |
| HALh | 20 | AGgcaagc | 2,000 |
| MNKi | 9 | AGgcaagt | 1,200 |
| FAAj | 16 | AGgcaagc | NA |
| FAA | 37 | AGgcaaga | NA |
| FAA | 41 | AGgcaggt | NA |
| CACNL1A3k | 32 | AGgcacgc | 1,000 |
| RYR1l | 57 | AGgcacgc | 596 |
| FE65m | 10 | AGgcacgg | 144 |
| XPGn | 3 | AGgcaaga | NA |
| Ftnbo | 13 | AGgcaaga | 3,100 |
| Relnp | 30 | AGgcaagt | 1,000 |
| TFIIS.oAq | 8 | AGgcaagt | NA |
| TGG1r | 1 | AGgcatgt | NA |
Intron 6 of the gene encoding human P120, a proliferating cell nucleolar antigen, and intron 7 of the gene encoding human CMP, a cartilage matrix protein (matrilin 1), were the first reported examples of introns with AT and AC at the intron ends, instead of GT and AG (38). Intron 6 of the gene encoding Rep-3, a DNA repair protein, and intron 2 of the gene encoding Prospero, a Drosophila melanogaster homeodomain transcription factor, also have AT-AC ends (28). In addition to their distinctive dinucleotide ends, these and other AT-AC introns have highly conserved 5′-splice-site and presumptive branch site 8-nucleotide sequence elements that are not present in the major class of introns, ATATCCTY and TCCTTRAY, respectively. On the basis of these sequence features, it was proposed that the minor U11 and U12 snRNAs, which have regions of complementarity to these elements, are required for splicing of AT-AC introns (28). Important aspects of this prediction were soon verified experimentally (29, 100), and two additional minor snRNAs involved in the novel pre-mRNA splicing pathway were discovered (101).
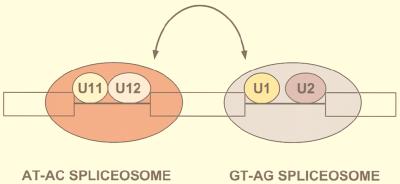
The AT-AC splicing pathway was originally named after the distinctive sequences of the intron ends (100). It was later found that a few introns with AT-AC ends are processed by the major pathway (120) and, conversely, that some introns with GT-AG ends are spliced by the minor pathway (18). Therefore, the name AT-AC no longer reflects the dinucleotide intron ends per se, but rather it refers to the minor pathway itself. An alternative designation for the two pathways—U2 dependent and U12 dependent—reflects their observed or expected requirements for one of the four snRNAs specific to each pathway (86).
DISTRIBUTION OF AT-AC INTRONS
AT-AC introns exist in a variety of organisms ranging from Arabidopsis thaliana to Drosophila, Xenopus laevis, and mammals (86, 102, 120). However, AT-AC introns are absent in budding yeast. There are no obvious structural relationships or common expression patterns among the genes or gene families that contain AT-AC introns. The lengths of known AT-AC introns range from less than 100 bases to more than 3,000 bases (120). The position of AT-AC introns is not conserved among unrelated genes. However, AT-AC introns are conserved phylogenetically and within gene families. For example, the last intron of the CMP gene is an AT-AC intron that is conserved in human, mouse, and chicken genes (4). Two genes with an AT-AC intron, mouse Rep-3 (a homologue of bacterial MutS) and XPG (a gene defective in xeroderma pigmentosum), are thought to be involved in DNA repair (120). The significance of the presence of AT-AC introns in DNA repair genes may be borne out as more sequences of DNA repair genes are determined. To date, three gene families have been noted to have AT-AC introns: the E2F transcription factor genes, the voltage-gated sodium and calcium channel α subunit genes (reference 120 and references therein), and the cartilage matrix protein (matrilin) family genes (4, 109). The voltage-gated ion channel genes are especially interesting because they have multiple minor introns; the conservation of minor introns in these genes is therefore reviewed in detail below.
IN VITRO AND IN VIVO SYSTEMS TO STUDY AT-AC PRE-MRNA SPLICING
Two different approaches have been used to study the mechanisms of AT-AC pre-mRNA splicing. The in vivo approach consists of transfecting wild-type or mutant minigenes containing an AT-AC intron and analyzing the splicing patterns after transient expression. Suppressor AT-AC snRNAs can be cotransfected with AT-AC intron mutants to test the functional significance of proposed base-pairing interactions between conserved intron elements and complementary regions in the snRNAs (29, 37, 45), as originally done for the conventional pathway (129).
The development of in vitro systems for AT-AC pre-mRNA splicing made it possible to begin to study the biochemical mechanisms of the reaction. To date, in vitro splicing conditions have been established for processing the AT-AC introns of pre-mRNAs from two different genes: the gene encoding P120 and that encoding SCN4A, the voltage-gated skeletal muscle sodium channel α subunit (100, 119). In vitro splicing of both P120 and SCN4A pre-mRNAs in HeLa cell nuclear extract results in generation of a lariat intermediate and release of the intron as a lariat. Therefore, AT-AC splicing occurs in two steps involving _trans_-esterification reactions similar to those of the major splicing pathway. The results are consistent in the two AT-AC in vitro splicing systems, but there is one major difference between them. Inactivation of U1 or U2 snRNAs in the nuclear extract is required to detect P120 AT-AC splicing (100; reviewed in reference 102). This observation led to the suggestion that U2 may compete with U12, which is about a hundred times less abundant (64), for binding to the AT-AC branch site (47). However, this is unlikely to be the case for SCN4A AT-AC splicing in vitro, which does not require inactivation of the major splicing pathway (119). In fact, SCN4A AT-AC splicing and cryptic splicing via conventional splice sites occur in the same reaction.
FOUR MINOR SNRNAS ARE REQUIRED FOR AT-AC PRE-MRNA SPLICING
AT-AC introns have unique and highly conserved 5′-splice-site and branch site elements, which are recognized by a unique set of minor snRNAs, U11, U12, U4atac, and U6atac. These snRNAs lack extensive sequence homology to the major snRNAs, but they appear to have related secondary structures, and more importantly, they play analogous roles in splice site recognition and perhaps in splicing catalysis. Despite the remarkable parallels, there are some significant differences between major and minor snRNAs. For example, U11 and U12 form a stable di-snRNP particle (111) that probably enters the AT-AC spliceosome as a single entity, whereas U1 and U2 are discrete snRNP mono-particles. Although the major U5 snRNA appears to be involved in both splicing pathways (100), this snRNA assembles onto the major spliceosome as part of a U4/U6 · U5 tri-snRNP particle, whereas an analogous U4atac/U6atac · U5 tri-snRNP particle has not yet been described. Notwithstanding the fact that the AT-AC and major spliceosomes have different snRNA constituents, the catalytic core of the AT-AC spliceosome is thought to resemble that of the major spliceosome (101; reviewed in references 9, 69, 70, and 102).
U11 and U12 snRNAs.
Human U11 and U12 are rare snRNAs that have Sm antigen binding sites but exhibit no sequence homology to other snRNAs (64). U11 and U12 snRNP particles presumably contain all the Sm core proteins. U12 interacts with a fraction of the more abundant U11 to form a di-snRNP complex (111). A 65-kDa protein of the U11/U12 complex, identified by virtue of its reactivity with a scleroderma patient antiserum has been described previously, although its sequence is not known (25). The predicted secondary structures of U11 and U12 snRNAs are similar to those of U1 and U2, respectively (64, 126). U11 and U12 localize in the nucleoplasm and are concentrated in coiled bodies and nuclear speckles, but they are excluded from nucleoli (58). This distribution is very similar to that of the major spliceosomal snRNAs U1 and U2 (reviewed in reference 89). U12 orthologues have been cloned from mouse, chicken, and frog (103, 126), species in which AT-AC introns are known to exist (reviewed in references 86, 102, and 120).
The role of U11 and U12 snRNAs in AT-AC splicing has been firmly established (29, 45, 100, 119, 125; reviewed in references 66, 70, and 102). U11 snRNA is present in in vitro-assembled P120 AT-AC spliceosomes (100) and can be cross-linked to the P120 AT-AC 5′ splice site (125). U11 also interacts with the P120 AT-AC 5′ splice site in vivo through base pairing (45). U12 snRNA is essential for AT-AC splicing in vitro and in vivo (29, 100, 119). U12 functions in the AT-AC splicing pathway by base pairing with the highly conserved branch site sequences (29, 100). Thus, U11 is analogous to U1 snRNA in the major pathway, whereas U12 is analogous to U2.
U11 and U12 snRNPs were also shown to be part of a negative regulator of splicing complex that inhibits splicing of Rous sarcoma virus pre-mRNA via the major pathway (26). This finding suggests a regulatory role for the U11/U12 di-snRNP particle in addition to its general role in AT-AC pre-mRNA splicing.
U4atac and U6atac snRNAs.
U4 and U6 snRNAs are not required for AT-AC splicing. U6 is highly conserved between yeasts and mammals and is thought to function at the catalytic core of the major spliceosome. This critical role of U6 in the conventional pathway suggested the existence of an analogous molecule for the AT-AC pathway (100). The spliceosomal U6 snRNA has a γ-monomethyl guanosine triphosphate (meGTP) cap structure. Several low-abundance snRNAs with a meGTP cap structure but otherwise structurally distinct from U6 were identified by immunoprecipitation with antibody specific for the meGTP cap structure (27). Using the same antibody, two novel minor snRNAs termed U4atac and U6atac were identified in affinity-purified AT-AC spliceosomes assembled in vitro on P120 pre-mRNA (101). U4atac has a trimethylguanosine cap but coprecipitated because of its tight association with U6atac, analogous to the interaction between U4 and U6. U4atac, like U4, is an Sm snRNA. U6atac, like U6, has a meGTP cap structure and terminates with a stretch of U residues, suggesting its transcription by RNA polymerase III. Both U4atac and U6atac were shown to be essential for in vitro splicing of the P120 AT-AC intron and subsequently also for the SCN4A AT-AC intron (101, 120).
The predicted secondary structure of the U4atac/U6atac snRNA complex is strikingly similar to that of the U4/U6 snRNA complex, although U4atac and U6atac exhibit only about 40% overall sequence homology to U4 and U6, respectively (101). In the central region of U6atac, the homology to U6 is much greater, about 80%, and this region includes sites that can be cross-linked to U12 with psoralen. U6atac, which has an AAGGAGA box at the region corresponding to the ACAGA box of U6, base pairs with the AT-AC 5′ splice site, replacing U11 (37, 125). The U6atac-U12 helix identified by psoralen cross-linking is analogous to a U6-U2 helix thought to be part of the major spliceosome active site, which strongly suggests that U6atac likewise functions at the catalytic core of the AT-AC spliceosome (101). U4atac appears to act as a chaperone for U6atac, similar to the role of U4 vis-à-vis U6 (101). U5 is apparently required for splicing of both AT-AC and GT-AG introns (100, 120). Whether U5 base pairs with AT-AC exon borders, as has been shown for conventional exon sequences (reviewed in reference 68), remains to be determined.
AT-AC SPLICE SITE RECOGNITION
The mechanisms by which conventional splice sites are selected with high fidelity in metazoan pre-mRNAs remain largely unknown (reviewed in reference 7). The short, degenerate splice site and branch site elements are clearly required for proper splice site recognition, but they are not sufficient (93). The arrangement, spacing, and sequence context of the splice sites probably also contribute to accurate splice site selection.
In the case of AT-AC introns, even though the 5′-splice-site and branch site sequences are highly conserved 8-nucleotide elements, they probably lack sufficient information to specify AT-AC splice site recognition. Genomic sequences contain many pairs of sequences that match the AT-AC 5′-splice-site and branch site elements, but probably only a few of these are authentic AT-AC intron elements. Therefore, the sequence complementarity between the minor snRNAs and the AT-AC 5′-splice-site and branch site sequences is not sufficient to accurately select AT-AC splice sites. As with conventional introns, additional mechanisms and/or auxiliary signals for AT-AC splicing may help identify the authentic splice sites. For example, the arrangement, spacing, and sequence context of AT-AC splice sites within the entire pre-mRNA, as well as the presence of intronic and exonic elements, are likely to contribute to accurate splice site selection.
Exon definition interactions between the AT-AC and major spliceosomes.
Most vertebrate genes have multiple introns, which are usually very large, whereas the exons are relatively small (30, 94). Sequences within large introns can match the degenerate splice site consensus elements, but they are not recognized by the spliceosome, at least in the presence of the wild-type splice sites. The exon definition model (reviewed in reference 6) proposes that in pre-mRNAs with large introns, the splicing machinery initially recognizes a pair of splice sites around a short exon and assembles on the exon; subsequently, neighboring exons are juxtaposed (80). Thus, the sequences within large introns need not be recognized. In lower eukaryotes, or in the case of small introns, a pair of splice sites at the ends of the short intron is directly recognized by the spliceosome, by an intron definition mechanism (97). Exon definition and intron definition are probably not mutually exclusive. Both types of interactions may occur simultaneously to facilitate the recognition of multiple splice sites.
The exon definition model predicts that in genes with multiple introns, exons and their flanking introns cannot both be large. Indeed, large introns are usually flanked by small exons and large exons are usually flanked by small introns (94, 128). Statistical analyses of the length of vertebrate internal regulated exons and primate internal exons showed that very large exons and very small exons are very rare (6, 91). Several lines of experimental evidence have given strong support to the exon definition model. First, mutation of a downstream 5′ splice site inhibits splicing of the upstream intron (97). Second, strengthening the downstream 5′ splice site increases the splicing efficiency of the upstream intron (49). Finally, joining a 5′ splice site to the end of the downstream exon increases the splicing efficiency of the upstream intron in vitro (48). Numerous cases of splice site mutations in vertebrate genes cause exon skipping rather than intron retention (46, 67). These phenomena are consistent with the exon definition model, although exon skipping can also be explained by _cis_-competition between splice sites or, in some cases, by instability of the retained-intron RNAs because of blocked mRNA export and/or nonsense-mediated mRNA decay. Evidence for the recognition of terminal exons also supports the exon definition model. Components that bind to the 5′ cap cooperate with the splicing machinery to facilitate the recognition of the first exon (52). Definition of the last exon involves cross talk between the splicing and polyadenylation machineries (71).
After exon definition, the correct exons must be ligated. The mechanisms responsible for the correct juxtaposition of exons are poorly understood. Members of the SR protein family, a group of essential pre-mRNA splicing factors with characteristic arginine-serine C-terminal repeats (RS domain) and one or two N-terminal RNA-recognition motifs, may play a bridging role for exon juxtaposition, since they can bind to exon sequences and also can interact with themselves (14, 92, 116). SR proteins can interact with U1-70K, a U1 snRNP-associated protein, and U2AF, a U2 auxiliary factor (116). It is thought that SR proteins can bridge partially assembled spliceosomes on neighboring exons through U1-70K and U2AF. Splicing factor 1 (SF1) may also play a bridging role for exon juxtaposition. SF1 interacts with both U2- and U1-associated factors (1, 78).
Because AT-AC introns always coexist with multiple major introns, the question arises whether exon definition interactions take place between the two different classes of introns. Indeed, in vitro splicing of the SCN4A AT-AC intron 2 is strongly stimulated when exon 3 is followed by the conventional 5′ splice site of intron 3. More importantly, the stimulatory effect is dependent on intact U1 snRNP (119, 121). Therefore, U1 bound at the downstream 5′ splice site interacts with the upstream AT-AC splicing machinery in vitro (Fig. 1). A 4-base deletion in the conventional 5′ splice site of SCN8A intron 3 results in complete skipping of exons 2 and 3 in vivo (43). This is probably due to disruption of exon definition interactions between the AT-AC intron 2 and the conventional intron 3. The fact that exon 2 does not join to exon 4 implies that an AT-AC 5′ splice site and a conventional 3′ splice site are incompatible, i.e., that the AT-AC 5′ spliceosomal components and the conventional 3′ spliceosomal components cannot be bridged by intron definition. Consistent with this idea, replacement of the SCN4A AT-AC intron 2 branch site element with a conventional branch site abrogates AT-AC splicing in vitro, resulting in stimulation of the conventional pathway via a pair of cryptic splice sites (118).
FIG. 1.
Exon definition interactions between consecutive minor and major introns. Components of the minor and major spliceosomes interact across the intervening exon. The spliceosomes are denoted by the ellipses. U11 and U12 are shown bound to the AT-AC 5′ splice site and branch site, respectively, and U1 and U2 are shown bound to the conventional 5′ splice site and branch site, respectively. The interaction across the exon requires U1 snRNA base pairing at the conventional 5′ splice site. Bound U1 snRNP probably interacts indirectly with U12 snRNP components, perhaps through bridging by SR proteins. U11 and U12 form a di-snRNP particle, although it is not known if their interactions are maintained in the spliceosome.
Exon definition interactions between the major and minor spliceosomes presumably involve indirect contacts between the major and minor snRNPs (Fig. 1). Whether exon definition can also take place between a downstream AT-AC intron and an upstream conventional intron is not known. The question also arises how the terminal exons are defined in the context of AT-AC introns, which are occasionally the first or the last intron. For example, the AT-AC intron in the calcium channel CACNL1A4 gene is the first intron (73), whereas the AT-AC introns in the matrilin family genes (4, 109) and GT335, a gene of unknown function (50), are the last intron of their respective genes. Therefore, it will be of interest to determine whether terminal exons can be defined by interactions between the AT-AC splicing machinery with 5′-cap recognition components or the polyadenylation machinery.
Purine-rich enhancers contribute to AT-AC splice site recognition.
In addition to splice site interactions that take place via intron or exon definition, splice site selection can also be facilitated by exonic and/or intronic sequences. Splicing enhancers are elements that contribute to the recognition of authentic splice sites. The actual prevalence of these elements is not yet known. Purine-rich sequences characteristic of most natural exonic splicing enhancers (ESEs) characterized to date are present in a wide variety of cellular and viral genes in metazoans. Purine-rich sequences have also been shown to modulate plant 5′ splice-site selection (61). In metazoans, purine-rich ESEs are recognized by members of the SR protein family. Purine-rich enhancers are usually composed of GAR repeats (R represents a purine nucleotide) but not runs of G or A (35, 98). However, G-rich repeats typical of small intron sequences in vertebrate genes may facilitate the recognition of splice sites in small introns (60). Non-purine-rich splicing enhancers can also stimulate splicing (16, 53, 104, 105).
Some exons downstream of known AT-AC introns have purine-rich sequences that closely resemble those present in natural enhancers of the major splicing pathway (Table 2), raising the question of whether purine-rich sequences can act as AT-AC splicing enhancers. Indeed, heterologous purine-rich sequences that function as enhancers in the conventional pathway also strongly stimulate AT-AC splicing when placed in the context of a downstream exon that naturally lacks such sequences (121). The purine-rich sequences that are present in many exons flanking AT-AC introns may be natural AT-AC splicing enhancers, or they may influence splicing of both conventional and AT-AC introns on either side. It will be interesting to determine whether non-purine-rich exonic enhancers or intronic enhancers can also function in the AT-AC splicing pathway. The finding that purine-rich enhancers can function in the AT-AC splicing pathway suggests that the enhancer-specific functions of SR proteins are relevant to the AT-AC splicing pathway. If this is the case, an important question is whether SR proteins interact with overlapping or with distinct components of the major and minor spliceosomes to mediate splicing enhancement.
TABLE 2.
Exonic purine-rich sequences present downstream of known AT-AC intronsa
| Gene product | Exon no. | Sequence |
|---|---|---|
| P120b | 7 | ...AGCGGGAGGAAGGGCGG...CTCAAGAAGGATCT... |
| CMPc | 8 | ...CCTGGAGAACAC... |
| GT335d | 7 | ...GGTGAGGAAGGTGCTGGAACTCACTGGAAAGTGA... |
| CDK5e | 10 | ...GACGAGGAACAA... |
| REP-3f | 7 | ...TACGAAGAAAAGGAGAACAT...GACAAAAAGAAGGGGAACCT... |
| HPSg | 16 | CTACCTGGAGGATTT...AACGAAAAGATGT...GGCAAGGGGCCC... |
| TFIIS.oAh | 7 | ...CCTGAGAAGAAATGT... |
In the major splicing pathway, both the downstream 5′ splice site and purine-rich enhancers contribute to the selection of the upstream splice site and appear to involve the action of SR proteins (reviewed in references 6, 7, 10, and 57). In addition, the U1 snRNP has also been implicated in enhancer function: U1 snRNA has been cross-linked to a purine-rich enhancer (112) and is present in an in vitro-assembled enhancer complex (90, 92). SR proteins facilitate U1 binding to 5′ splice sites (21, 44, 127), and it follows that SR proteins bound to purine-rich enhancers may also recruit U1 (reviewed in references 10, 57, and 108). However, intact U1 snRNP is not required for the stimulation of AT-AC splicing by purine-rich enhancers in vitro (121). Therefore, enhancer function and exon definition are mechanistically different, at least in terms of a requirement for U1.
In the major pathway, ESEs and the downstream 5′ splice site facilitate recognition of the upstream 3′ splice site by specific interactions with SR proteins. These proteins apparently interact with the essential splicing factor U2AF, facilitating its binding to the 3′-splice-site polypyrimidine tract, while U2AF in turn promotes binding of U2 to the branch site (31, 130). The 3′ region of AT-AC introns differs from that of conventional introns: the former lack a polypyrimidine tract, and the branch site is much closer to the 3′ splice junction. These differences suggest that the AT-AC spliceosomal components involved in recognition of the 3′ splice site are different from those of the conventional spliceosome. Surprisingly, despite these differences, both a downstream 5′ splice site and purine-rich enhancers stimulate AT-AC splicing, suggesting that both contribute to AT-AC splice-site selection, as they do in the major splicing pathway. Although in the AT-AC splicing pathway both exonic enhancers and a downstream conventional 5′ splice site are also expected to interact with spliceosomal components at the upstream 3′ splice site, some of these components may be unique to the AT-AC splicing pathway.
A DIVERGENT SUBCLASS OF AT-AC INTRONS
In vitro studies showed that splicing of the SCN4A intron 2 occurs by the AT-AC splicing pathway, and this is probably also the case for the homologous introns in the voltage-gated sodium and calcium channel genes (119, 120). Interestingly, the SCN4A gene has another unusual intron, intron 21, which has 5′-AT and AC-3′ ends (24, 59, 120), as does the corresponding intron 25 of the SCN5A gene (110, 120). However, the 5′-splice-site and branch site elements of these two introns do not match the highly conserved AT-AC 5′-splice-site and branch site consensus sequences, suggesting that these divergent introns belong to a distinct subclass of AT-AC introns (119). Indeed, in vitro splicing of these two sodium channel introns, termed AT-AC II, requires the major U1, U2, U4, U5, and U6 spliceosomal snRNAs, rather than the minor AT-AC snRNAs (120). Other AT-AC II introns have previously been described in the chicken parvalbumin gene and in xylanase genes of several filamentous fungi (120).
The vast majority of introns have GT and AG boundaries at the intron 5′ and 3′ ends, respectively. The importance of the intron ends is underscored not only by in vitro and in vivo mutational analyses (3) but also by the fact that splice site mutations impair gene expression and cause numerous human genetic diseases (46, 67). On the other hand, previous mutational analyses showed that certain splice site mutations are compatible with accurate splicing. For example, in the yeast actin intron and a human tropomyosin intron, mutation of G to A at the first position or G to C at the last position compromises splicing; however, the double mutation at both ends of these introns allows accurate splicing in vivo, albeit less efficiently (13, 74, 85). The double mutations generate 5′-AT and AC-3′ intron boundaries. Although these observations preceded the discovery of a unique AT-AC splicing pathway, a possible explanation of these results is that the double-mutant pre-mRNAs were processed by the minor pathway. This appears unlikely, because the double-mutant intron sequences do not match the highly conserved AT-AC 5′-splice-site and branch site consensus (66). Although the snRNA requirements for splicing of the mutant introns have not been determined, it is likely that they are processed by the major pathway, by analogy to the natural AT-AC II introns, which require the major snRNAs (120). The mutational results were interpreted to suggest a non-Watson-Crick interaction between the first and last bases of the introns. Analysis of the splicing of introns with inosine inserted at the intron ends supports this model (85, 99). The non-Watson-Crick interaction between the intron ends probably exists in the natural AT-AC II introns and probably also exists in all AT-AC introns. On the other hand, the observed lack of specificity in selection of the last nucleotide of a yeast intron argues against a direct interaction between the first and last bases of introns (55).
Although the splice site sequences and the positions of the SCN4A AT-AC II intron 21 and SCN5A AT-AC II intron 25 are conserved, the lengths of these two introns are different. The longer SCN4A AT-AC II intron 21 contains an Alu repeat insertion at nucleotides 358 to 660 (numbering as per GenBank entry AF007782) (117). Alu sequences are repetitive elements that are unique to primates and are thought to be derived from the 7SL RNA (reviewed in reference 62). The functions of Alu sequences are unclear, although in some cases they can influence gene expression. Alu sequences contain several regions that differ from either 5′ or 3′ splice site consensus elements by only one or two nucleotides. When Alu sequences are present within conventional introns, they can have dramatic effects on splicing (reviewed in reference 56). Point mutations can activate cryptic splice sites in intronic Alu elements and result in abnormal protein formation and clinical disease, e.g., Alport syndrome and ornithine delta-aminotransferase deficiency (41, 63). Mutations in the SCN4A gene cause hyperkalemic periodic paralysis and paramyotonia congenita (reviewed in reference 32). Whether there are natural mutations of this gene that involve activation of cryptic splice sites within the Alu insertion in intron 21 or whether this Alu insertion has any functional consequences remains to be seen.
U12-TYPE GT-AG INTRONS
AT-AC II introns have 5′-AT and AC-3′ boundaries, yet their splicing requires the major spliceosomal snRNAs (120). Conversely, some introns that have 5′-GT and AC-3′ boundaries are spliced by the minor spliceosomal snRNAs (18). With the exception of the first and last nucleotides, which are both G, the sequences of these introns match the AT-AC splice site and branch site consensus elements (Tables 3 and 4). This subclass of GT-AG introns has been termed U12-type GT-AG introns (86). Recent compilations showed that U12-type GT-AG introns are more prevalent than AT-AC introns (18, 86). Additional examples of this type of intron are shown in Tables 3 and 4. Interestingly, XPG- and CDK5-encoding genes contain both an AT-AC intron and a U12-type GT-AG intron (54, 72). Likewise, the members of the voltage-gated ion channel α subunit gene family have several unconventional introns (see below). Intron 2 of the gene encoding human CACNLB3, the voltage-gated calcium channel β subunit, also belongs to the U12-type GT-AG intron class.
TABLE 3.
Compilation of U12-type GT-AG intronsa
| Gene product | Intron no. | 5′ splice site | Presumptive branch site | 3′ splice site | Distance (nt) | Size (nt) | |
|---|---|---|---|---|---|---|---|
| MDCc | 11 | AGgtatcctc | ... | ctcac | ccctcagGG | 9 | 109 |
| HALd | 3 | CTgtatcctt | ... | NAb | ctggatacagAT | NA | 100 |
| HAL | 6 | AGgtatcttt | ... | NA | tttattctagTT | NA | 100 |
| HAL | 15 | CTgtatcttt | ... | NA | cttttgaaagAT | NA | 3,000 |
| MNKe | 11 | AGgtatttat | ... | atgttaac | ttatatccagTG | 12 | 1,000 |
| BMP1f | 15 | AGgtatcc | ... | NA | ttctgttgctccagTC | NA | NA |
| BMP1 | 15 | ... | NA | ctctctcgtttcagAA | NA | NA | |
| BMP1 | 15 | ... | NA | ttgctcccctgcagAG | NA | NA | |
| CACNL1A1g | 15 | CTgtatcctt | ... | NA | acacaaacagAT | NA | 1,600 |
| CACNL1A2h | 16 | CTgtatcctt | ... | cccttaaa | aagttgaaattagAT | 15 | 3,400 |
| CACNL1A3i | 13 | CTgtatcctc | ... | tccttagc | taaacccgctcagAC | 15 | 900 |
| CACNL1A4j | 16 | CTgtatcctt | ... | tcctgact | cagacatttgcagAC | 15 | 155 |
| CACNLB3k | 2 | AGgtatactt | ... | gcactaat | gggcaaattctccagCA | 17 | 216 |
| MSH5l | 6 | TGgtatctcc | ... | ccctcaaa | tagGT | 5 | 733 |
| Relnm | 27 | AGgtatct | ... | tccttcac | tagCT | 5 | 2,000 |
| SCN10An | 8 | AGgtatctt | ... | cccttgaa | tctccagAC | 9 | 300 |
| XPGo | 1 | GGgtatcctt | ... | tcctttac | tggttcccccagAT | 14 | NA |
| CDK5p | 6 | GGgtatctgt | ... | NA | ctcccctcagAA | NA | 470 |
TABLE 4.
Minor intron conservation in voltage-gated sodium and calcium channel α subunit genesa
| Gene product | Intron no. | 5′ splice site | Presumptive branch site | 3′ splice site | Distance (nt) | Size (nt) | |
|---|---|---|---|---|---|---|---|
| SCN4Ac | 2 | GCatatcctg | ... | tccttgac | cctgccccacGC | 12 | 126 |
| SCN5Ad | 3 | TCatatcc | ... | NAb | cccacgcacGC | NA | NA |
| SCN8Ae | 2 | TCatatcctt | ... | cccttaac | tcctctctacAG | 12 | NA |
| SCN10Af | 2 | TCgtgtcct | ... | tccttaac | atggacctcacagCT | 15 | 4200 |
| CACNL1A1g | 2 | AAatatcctt | ... | tccttgac | tccctttctcagAC | 14 | 5000 |
| CACNL1A2h | 2 | AAgtatcctt | ... | accttaac | acattttttcagAC | 14 | 4800 |
| CACNL1A3i | 1 | AAgtatcctt | ... | tccttaac | cctgctccagGC | 12 | 600 |
| CACNL1A4j | 1 | CCatatcctt | ... | tccttaat | tccccaatacTC | 12 | NA |
OTHER NATURAL INTRON BOUNDARIES
AT-AA introns.
Previous studies showed that a G-to-A mutation at the last position of a major class intron can partially suppress a G-to-A mutation at the first position of the intron in vivo (11, 74). To date, three natural AT-AA introns are known: intron 6 of the gene encoding the DNA excision repair protein hMSH3 (113); intron 7 of the gene encoding Arabidopsis AtG5 (115); and intron 3 of the gene encoding pig succinyl-coenzyme A (CoA) synthetase (82) (Table 5). Intron 6 of the gene encoding hMSH3 and intron 7 of the gene encoding AtG5 have the AT-AC 5′-splice-site consensus. Intron 6 of the gene encoding hMSH3 also has the AT-AC branch site consensus and is the homologous intron of the AT-AC intron 6 of the Rep-3 gene. Therefore, this intron is almost certainly processed via the AT-AC pathway. The corresponding intron 6 of the related hMSH5 gene is a U12-type GT-AG intron (Table 3). Interestingly, in alternative splicing of hMSH2, another related DNA repair gene, intron 12 was reported to have unusual TA-TT ends (65). Although the AT-AA intron 7 of the gene encoding AtG5 may be processed by the AT-AC pathway, this assumption needs to be verified experimentally, since the intron lacks a good match to the AT-AC branch site consensus. Intron 3 of the gene encoding pig succinyl-CoA synthetase is alternatively spliced and lacks the AT-AC 5′-splice-site and branch site consensus. Therefore, splicing of this intron likely requires the major snRNAs.
TABLE 5.
Compilation of known AT-AA intronsa
| Gene product | Intron no. | 5′ splice site | Presumptive branch site | 3′ splice site | Distance (nt) | Size (nt) | |
|---|---|---|---|---|---|---|---|
| hMSH3c | 6 | AGatatcctt | ... | tctttaat | tattattaaAT | 11 | 2,000 |
| AtG5d | 7 | AAatatcctt | ... | atattaac | caaggcttaaGT | 12 | 138 |
| SCSe | 3 | TGataagt | ... | tttttgaa | ttctccttctttaaGG | 16 | NAb |
AT-AG introns.
Mutation of the first nucleotide from G to A of a major-class intron inhibits the second step of splicing in vitro (13, 74, 85). Many natural mutations giving rise to various genetic diseases consist of G-to-A mutation at the first position of an intron, with the consequent impairment of gene expression (46, 67). For example, this type of mutation in intron 1 of the human β-globin gene prevents normal splicing, activates three cryptic 5′ splice sites, and causes thalassemia (106); in intron 5 of the human adenosine deaminase gene, it results in severe immunodeficiency (84); in intron 5 or intron 18 of the human myosin VIIA gene, it causes Usher syndrome type IB (2). These in vitro and in vivo observations suggested that 5′-AT and AG-3′ boundaries within the same introns are incompatible. This is likely to be true in the context of the major splicing pathway, so the few natural examples of AT-AG introns are likely to be processed via the minor splicing pathway.
Three natural introns have 5′-AT and AG-3′ ends: intron 38 of the myosin VIIa gene (40, 51), which codes for a long-tailed unconventional myosin; intron 2 of CACNL1A1 (87); and intron 24 of SCN10A (88). Intron 38 of the myosin VIIa gene has the 5′-splice-site and presumptive branch site sequences ATATCCGT and TCCTTGAC, respectively, as independently reported by two different groups (40, 51). These sequences are close matches to the AT-AC consensus elements. The introns corresponding to intron 2 of CACNL1A1 in other voltage-gated calcium channel α subunit genes are either of the AT-AC class or of the U12-type GT-AG class. Thus, splicing of all three types of introns presumably requires the minor snRNAs, although this assumption needs to be confirmed experimentally. Splicing via the minor pathway would account for the tolerance to 5′-AT and AG-3′ intron ends (see below).
DIFFERENTIAL TOLERANCE TO MUTATION OF THE LAST INTRON NUCLEOTIDE IN AT-AC AND CONVENTIONAL SPLICING
Most natural AT-AG and AT-AA introns have close matches to the highly conserved AT-AC 5′-splice-site and branch site consensus sequences. The distances between the branch site and the 3′-splice junction are all very short. Therefore, it appears that the AT-AC spliceosome tolerates changes in the last intron nucleotide, which can be either C, A, or G. Indeed, a recent in vivo mutational analysis of the last nucleotide of the AT-AC intron in the P120 gene showed that although C-to-G mutation results in activation of a pair of conventional cryptic splice sites, accurate splicing via the AT-AC pathway can still occur (18).
The 5′-AT and AG-3′ boundaries of the CACNL1A1 intron 2 may represent an evolutionary transition between the AT-AC introns in the voltage-gated sodium channel α subunit genes and the U12-type GT-AG introns within the homologous domain I of the voltage-gated calcium channel α subunit genes. Intron 2 of mouse SCN10A, which has a GTGTCC 5′ splice site and a canonical AT-AC branch site (Table 4), may represent a further evolutionary transition from a U12-type GT-AG intron to a conventional intron. The 5′-AT and AA-3′ boundaries of hMSH3 intron 6 represent a drift from the corresponding AT-AC intron 6 of the homologous mouse Rep-3 gene. The 5′-GT and AG-3′ boundaries of hMSH5 represent a further drift. The major spliceosome has a more stringent sequence requirement for the last intron nucleotide, which can only be a G, unless the first nucleotide is simultaneously mutated. Conversely, in AT-AC II introns, the last nucleotide presumably has to be a C, unless the first nucleotide changes simultaneously. It is probably difficult for an AT-AC II intron to evolve into a conventional intron, or vice versa, because this would require simultaneous mutations at both ends of the intron. In contrast, AT-AC introns may more easily evolve gradually into conventional introns. The last nucleotide, C, can be first mutated to G, then the first nucleotide, A, can be mutated to G, and splicing would still be catalyzed by the minor spliceosome. Then the +5 position of the 5′ splice site can change from C to G. This last mutation may be sufficient for switching from the minor splicing pathway to the major splicing pathway and would likely constitute an irreversible switch, since AT-AC introns have highly conserved 5′-splice-site and branch site elements, whereas conventional introns have much more degenerate elements.
COMMITMENT TO A SPECIFIC SPLICING PATHWAY
The dinucleotides at the intron ends are not responsible for commitment of a pre-mRNA to the major or the minor splicing pathway (18, 120). Likewise, the highly conserved AT-AC branch site consensus sequence (UCCUURAY) recognized by U12 snRNA is probably not the major determinant of commitment to the minor pathway, since it also matches the degenerate consensus for conventional branch sites (YNYURAY), and therefore it should also be recognized by the abundant U2 snRNA. Intron 7 of the xylanase xylP gene in the filamentous fungus Penicillium chrysogenum has a sequence identical to the AT-AC branch site consensus, but it is probably processed via the major splicing pathway (120). It is likely that the highly conserved AT-AC 5′ splice site, which is recognized by U11 and U6atac snRNAs, is the major determinant of commitment to the AT-AC splicing pathway. An AT-AC branch site sequence is of course also required but is probably compatible with either pathway. The lack of an extensive polypyrimidine tract and the short distance between the branch site and the 3′ splice site in AT-AC introns probably also contribute to their specific commitment to the AT-AC splicing pathway.
MULTIPLE MINOR INTRONS IN VOLTAGE-GATED ION CHANNEL GENES
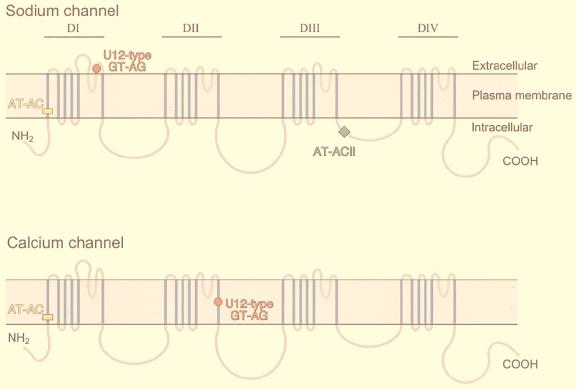
The voltage-gated sodium channel and calcium channel α subunit genes belong to the voltage-gated ion channel gene superfamily. Mutations in these genes cause numerous neuromuscular and neurological diseases (reviewed in reference 19). The genes have four internal homologous domains (I to IV) that are thought to arise from two rounds of duplication from a single ancestral gene (Fig. 2) (12, 42). Interestingly, these genes have two or sometimes three nonconsensus introns (Fig. 2; Tables 3, 4, and 6). Multiple sequence alignments of the different family members show that these introns interrupt the genes at exactly homologous positions of the coding sequence (Fig. 3).
FIG. 2.
Topology of voltage-gated ion channel proteins and positions of the unusual introns. The diagram shows the proposed topology of the voltage-gated sodium and calcium channel α subunit protein families (12, 42). Domain repeats I through IV (DI through DIV) are indicated at the top. Within each domain repeat, the six membrane-spanning segments are shown in blue, and the connecting loops are shown in gray. The positions of unusual introns that interrupt the coding sequences of the corresponding genes are indicated. For some of these introns, the first and last nucleotides are not invariant among different family members (Tables 4 and 6). The conservation of the position of the AT-AC introns (yellow rectangles) in domain I of the sodium and calcium channel genes is indicative of the presence of this intron prior to the divergence of these two gene families. The U12-type GT-AG introns (red circles) are present at different positions of the sodium and calcium channel genes. The AT-AC II intron (green square) is only present in the sodium channel genes. Some family members have lost one or more of these introns or have conventional introns at the same position. Each type of intron interrupts a different part of the domain repeat coding sequence, and hence each intron at the four indicated locations in the voltage-gated ion channel genes probably arose independently.
TABLE 6.
AT-AC II intron conservation in voltage-gated sodium channel α subunit genesa
| Gene product | Intron no. | 5′ splice site | Presumptive branch site | 3′ splice site | Distance (nt) | Size (nt) | ||
|---|---|---|---|---|---|---|---|---|
| SCN4Ac | 21 | AGatgagtat | ... | acctgac | ... | ccactatacTT | 21 | 822 |
| SCN5Ad | 25 | AGatacgtgg | ... | ctctgag | ... | tctttgcacTT | 37 | 680 |
| SCN8Ae | 21 | AGataggtct | ... | NAb | cctcctttacacTT | NA | NA | |
| SCN10Af | 24 | AGataagtg | ... | cgttaat | tcctcccccctagTT | 15 | 900 |
FIG. 3.
Conserved positions of unusual introns in voltage-gated ion channels. Multiple protein sequence alignments of the relevant regions of the voltage-gated sodium and calcium channel α subunits are shown. The complete amino acid sequences encoded by the flanking exons were aligned by using the Genetics Computer Group Pileup program, with a gap creation penalty of 12 and a gap extension penalty of 4 (23). The positions of the unusual introns in the corresponding genes are indicated by the vertical black bar. Amino acid identities in more than half of the sequences are indicated by gray shading. Complete exon sequences are shown, except where indicated. For each alignment, the intron position is also conserved at the nucleotide level (117). (A) Conserved positions of the minor AT-AC introns in eight ion channel genes. Translations of the 5′ portions of the calcium channel upstream exons are not shown in the alignment. The human SCN5A sequence is from reference 110; human SCN4A, mouse SCN8A, mouse SCN10A, and human CACNL1A1, CACNL1A2, CACNL1A3, and CACNL1A4 have GenBank accession no. L04216, U59964, Y09108, AJ224873, D43706, U30666, and X99897, respectively. (B) Conserved positions of the minor U12-type GT-AG introns in four calcium channel genes. Human CACNL1A1, CACNL1A2, CACNL1A3, and CACNL1A4 sequences have GenBank accession no. AJ224873, D43718, U30677, and X99897, respectively. (C) Conserved positions of the AT-AC II introns in four sodium channel genes. Human SCN5A and SCN8A sequences are from references 110 and 77, respectively; human SCN4A and mouse SCN10A sequences have GenBank accession no. L04233 and Y09108, respectively.
SCN4A, SCN5A, and SCN8A, the voltage-gated sodium channel α subunit in skeletal muscle, cardiac muscle, and brain and spinal cord, respectively, have two rare introns: an AT-AC intron and an AT-AC II intron (43, 77, 120; Fig. 2). The AT-AC intron interrupts the coding sequence of internal domain I, while the AT-AC II intron is located between domains III and IV. Interestingly, the corresponding AT-AC intron 2 of the mouse gene encoding SCN10A, the sensory neuron voltage-gated sodium channel α subunit, has the sequence GTGTCCT at the 5′ splice site and AG as the 3′-splice-site dinucleotide (Table 4) (88). The voltage-gated calcium channel α subunit genes also have an unusual intron at precisely the same position within domain repeat I (Fig. 2 and 3A). However, these introns have different boundaries (Table 4): intron 2 of the gene encoding CACNL1A1, the fibroblast L-type voltage-gated calcium channel α subunit, has 5′-AT and AG-3′ boundaries; intron 1 of the gene encoding CACNL1A4, the brain-specific P/Q-type voltage-gated calcium channel α subunit, has 5′-AT and AC-3′ boundaries; and intron 2 of the gene encoding CACNL1A2, the pancreatic voltage-gated calcium channel α subunit, and intron 1 of the gene encoding CACNL1A3, the skeletal muscle voltage-gated calcium channel α subunit, have 5′-GT and AG-3′ boundaries. All eight introns have the highly conserved AT-AC branch site element. The same kind of intron is expected to be present in other voltage-gated sodium and calcium channel α subunit genes and to be processed via the AT-AC splicing pathway (120). Splicing of an intron of the ADP ribose polymerase gene, which has the features of U12-type GT-AG introns, requires the minor U12 and U6atac snRNAs (18). It is therefore very likely that the GT-AG introns in CACNL1A2 (CACN4) and CACNL1A3 are also processed via the minor pathway.
Interestingly, the CACNL1A2 and CACNL1A3 genes have a second minor intron—a U12-type GT-AG intron—which falls within domain repeat II (Table 3; Fig. 2). This type of intron also has a precisely conserved position in all four known calcium channel α subunit gene sequences (Fig. 3B). These observations led to the prediction that analogous U12-type GT-AG introns may exist in other voltage-gated calcium channel α subunit genes. Alignment of the sequences of domain repeats I and II shows that the two minor introns do not interrupt a homologous position. The different location of these introns within their respective domain repeats suggests that they arose independently.
The SCN10A gene is also unusual in having three rare introns out of a total of 26 introns (Fig. 2): the U12-type GT-AG introns 2 and 8 (Tables 3 and 4) and the AT-AC II intron 24 (Table 6). The SCN4A intron 8 and SCN5A intron 9, which correspond to SCN10A intron 8, are conventional GT-AG introns. The voltage-gated calcium channel α subunit genes lack an intron at the corresponding position. This sequence comparison strongly suggests a shift from a U12-type GT-AG intron to a conventional GT-AG intron by mutational drift after the divergence of sodium and calcium channels from a common ancestral gene.
The SCN4A, SCN5A, SCN8A, and SCN10A genes are paralogues and belong to the voltage-gated sodium channel α subunit gene family. The genomic organization of these four genes is very similar (77, 88, 110). SCN4A, SCN5A, and SCN8A have an AT-AC II intron at a homologous position (Table 6). Interestingly, the corresponding intron 24 of SCN10A has 5′-AT and AG-3′ boundaries (Table 6) (88). All four introns interrupt a homologous position of the coding sequence of their respective gene (Fig. 3C). However, the voltage-gated calcium channel α subunit genes lack an intron at the homologous position (Fig. 2). Therefore, similar introns may exist in other members of the voltage-gated sodium channel α subunit genes but not in the voltage-gated calcium channel α subunit genes. Because splicing of the corresponding AT-AC II introns of SCN4A and SCN5A requires the major snRNAs, this is also expected to be the case for SCN8A intron 21 and SCN10A intron 24. The 5′-AT and AG-3′ boundaries of SCN10A intron 24 may represent an evolutionary transition between the corresponding AT-AC II introns and the major GT-AG introns.
In summary, the voltage-gated sodium and calcium α subunit genes, which are derived from a common ancestral gene, show an unusually high frequency of noncanonical introns. The distribution of introns among the two gene families and individual genes, the position of the introns within domain repeats of the coding sequence, and the patterns of sequence divergence at the intron ends provide interesting information about the evolutionary history of minor introns.
CONCLUSION
Although the first examples of AT-AC intron sequences were discovered only recently (28, 38, 100), a considerable amount of information about the AT-AC pre-mRNA splicing pathway has already been obtained. It is now known that AT-AC introns are widespread in higher eukaryotes (86, 102, 120). The removal of AT-AC introns from pre-mRNA requires the four minor U11, U12, U4atac, and U6atac snRNAs, which play roles analogous to those of U1, U2, U4, and U6 snRNAs in the major splicing pathway, and the major U5 snRNA, which is the only snRNA component shared with the major spliceosome (reviewed in references 70 and 102). Both exon definition interactions and purine-rich exonic enhancers stimulate AT-AC splicing in vitro (119, 121). Therefore, they may contribute to AT-AC splice site recognition in vivo.
Introns exist in the majority of eukaryotic cellular or viral genes. The mechanisms of splicing, including both catalysis and splice site selection, and the regulation of splicing are incompletely understood at present. However, the identification and characterization of some of the many protein factors involved in splicing continue to provide important clues about splicing and splice site selection mechanisms. The unexpected discovery of the AT-AC splicing pathway provides additional challenges and opportunities for understanding splicing mechanisms and specificity. Whereas the snRNAs involved in AT-AC splicing have been identified, no information is currently available about snRNP and non-snRNP proteins involved in this pathway. It will be especially interesting to determine whether some protein components are shared between the two pathways, and/or whether novel components—which may or may not resemble splicing factors in the conventional pathway—are uniquely required for AT-AC splicing. Determining which components are involved in interactions between the two pathways, such as in exon definition, should also be an important priority. Identification and characterization of these components should provide interesting insights into the mechanisms, regulation, and evolution of pre-mRNA splicing.
ADDENDUM
Shortly after this review was written, Burge et al. provided an extensive classification of U12-type and U2-type introns on the basis of a statistical analysis of 5′-splice-site and branch site sequences (9a). They describe instances of apparent intron conversion from U12 type to U2 type during evolution by examining introns of homologous or paralogous genes, and they discuss models for the evolution of the major and minor spliceosomes.
Comparison of the cDNA and genomic sequences (GenBank no. AF051782, AC005366, and AC005368) of HDIA1 (human diaphanous 1), a nonsyndromic deafness susceptibility gene, reveals the presence of two AT-AC introns and one GC-AG intron (118).
ACKNOWLEDGMENTS
We thank M. Hastings, M. Murray, and B. Graveley for helpful comments on the manuscript and M. Meisler for sharing unpublished sequence information.
The work on AT-AC pre-mRNA splicing in our laboratory is supported by grant GM42699 from the NIH to A.R.K.
REFERENCES
- 1.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 2.Adato A, Weil D, Kalinski H, Pel-Or Y, Ayadi H, Petit C, Korostishevsky M, Bonne-Tamir B. Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am J Hum Genet. 1997;61:813–821. doi: 10.1086/514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebi M, Hornig H, Weissmann C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- 4.Aszodi A, Beier D R, Hiripi L, Bosze Z, Fassler R. Sequence, structure and chromosomal localization of Crtm gene encoding mouse cartilage matrix protein and its exclusion as a candidate for murine achondroplasia. Matrix Biol. 1998;16:563–573. doi: 10.1016/s0945-053x(98)90067-1. [DOI] [PubMed] [Google Scholar]
- 5.Bao Y, Dawson T L, Jr, Chen Y T. Human glycogen debranching enzyme gene (AGL): complete structural organization and characterization of the 5′ flanking region. Genomics. 1996;38:155–165. doi: 10.1006/geno.1996.0611. [DOI] [PubMed] [Google Scholar]
- 6.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 7.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 8.Breathnach R, Benoist C, O’Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci USA. 1978;75:4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burge, C. B., T. H. Tuschl, and P. A. Sharp. Splicing of precursors to messenger RNAs by the spliceosome, p. 561–584. In R. F. Gesteland, and J. F. Atkins (ed.), The RNA world II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9a.Burge C B, Padgett R A, Sharp P A. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 10.Cáceres J F, Krainer A R. Mammalian pre-mRNA splicing factors. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: IRL Press; 1997. pp. 174–212. [Google Scholar]
- 11.Carothers A M, Urlaub G, Grunberger D, Chasin L A. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1993;13:5085–5098. doi: 10.1128/mcb.13.8.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catterall W A. Structure and function of voltage-sensitive ion channels. Science. 1988;242:50–61. doi: 10.1126/science.2459775. [DOI] [PubMed] [Google Scholar]
- 13.Chanfreau G, Legrain P, Dujon B, Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3′ splice site selection in S. cerevisiae. Nucleic Acids Res. 1994;22:1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew, S., H.-X. Liu, A. Mayeda, and A. R. Krainer. An exonic splicing enhancer facilitates the second catalytic step of pre-mRNA splicing. Submitted for publication.
- 15.Cho C, Turner L, Primakoff P, Myles D G. Genomic organization of the mouse fertilin beta gene that encodes an ADAM family protein active in sperm-egg fusion. Dev Genet. 1997;20:320–328. doi: 10.1002/(SICI)1520-6408(1997)20:4<320::AID-DVG3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dierick H A, Ambrosini L, Spencer J, Glover T W, Mercer J F. Molecular structure of the Menkes disease gene (ATP7A) Genomics. 1995;28:462–469. doi: 10.1006/geno.1995.1175. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich R C, Incorvaia R, Padgett R A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 19.Doyle J L, Stubbs L. Ataxia, arrhythmia and ion-channel gene defects. Trends Genet. 1998;14:92–98. doi: 10.1016/s0168-9525(97)01370-x. [DOI] [PubMed] [Google Scholar]
- 20.Durkin M E, Gautam M, Loechel F, Sanes J R, Merlie J P, Albrechtsen R, Wewer U M. Structural organization of the human and mouse laminin beta2 chain genes, and alternative splicing at the 5′ end of the human transcript. J Biol Chem. 1996;271:13407–13416. doi: 10.1074/jbc.271.23.13407. [DOI] [PubMed] [Google Scholar]
- 21.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimaki K, Yamazaki T, Taniwaki M, Ichinose A. The gene for human protein Z is localized to chromosome 13 at band q34 and is coded by eight regular exons and one alternative exon. Biochemistry. 1998;37:6838–6846. doi: 10.1021/bi972002a. [DOI] [PubMed] [Google Scholar]
- 23.Genetics Computer Group. Program manual for the Wisconsin Package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 24.George A L, Jr, Iyer G S, Kleinfield R, Kallen R G, Barchi R L. Genomic organization of the human skeletal muscle sodium channel gene. Genomics. 1993;15:598–606. doi: 10.1006/geno.1993.1113. [DOI] [PubMed] [Google Scholar]
- 25.Gilliam A C, Steitz J A. Rare scleroderma autoantibodies to the U11 small nuclear ribonucleoprotein and to the trimethylguanosine cap of U small nuclear RNAs. Proc Natl Acad Sci USA. 1993;90:6781–6785. doi: 10.1073/pnas.90.14.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gontarek R R, McNally M T, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, Busch R K, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J Biol Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- 28.Hall S L, Padgett R A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 29.Hall S L, Padgett R A. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 30.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman E P. Voltage-gated ion channelopathies: inherited disorders caused by abnormal sodium, chloride, and calcium regulation in skeletal muscle. Annu Rev Med. 1995;46:431–441. doi: 10.1146/annurev.med.46.1.431. [DOI] [PubMed] [Google Scholar]
- 33.Hogan K, Gregg R G, Powers P A. The structure of the gene encoding the human skeletal muscle alpha 1 subunit of the dihydropyridine-sensitive L-type calcium channel (CACNL1A3) Genomics. 1996;31:392–394. doi: 10.1006/geno.1996.0066. [DOI] [PubMed] [Google Scholar]
- 34.Hu Q, Kukull W A, Bressler S L, Gray M D, Cam J A, Larson E B, Martin G M, Deeb S S. The human FE65 gene: genomic structure and an intronic biallelic polymorphism associated with sporadic dementia of the Alzheimer type. Hum Genet. 1998;103:295–303. doi: 10.1007/s004390050820. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey M B, Bryan J, Cooper T A, Berget S M. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ianzano L, D’Apolito M, Centra M, Savino M, Levran O, Auerbach A D, Cleton-Jansen A M, Doggett N A, Pronk J C, Tipping A J, Gibson R A, Mathew C G, Whitmore S A, Apostolou S, Callen D F, Zelante L, Savoia A. The genomic organization of the Fanconi anemia group A (FAA) gene. Genomics. 1997;41:309–314. doi: 10.1006/geno.1997.4675. [DOI] [PubMed] [Google Scholar]
- 37.Incorvaia R, Padgett R A. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA. 1998;4:709–718. doi: 10.1017/s1355838298980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson I J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991;19:3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katagiri T, Harada Y, Emi M, Nakamura Y. Human metalloprotease/disintegrin-like (MDC) gene: exon-intron organization and alternative splicing. Cytogenet Cell Genet. 1995;68:39–44. doi: 10.1159/000133884. [DOI] [PubMed] [Google Scholar]
- 40.Kelley P M, Weston M D, Chen Z Y, Orten D J, Hasson T, Overbeck L D, Pinnt J, Talmadge C B, Ing P, Mooseker M S, Corey D, Sumegi J, Kimberling W J. The genomic structure of the gene defective in Usher syndrome type Ib (MYO7A) Genomics. 1997;40:73–79. doi: 10.1006/geno.1996.4545. [DOI] [PubMed] [Google Scholar]
- 41.Knebelmann B, Forestier L, Drouot L, Quinones S, Chuet C, Benessy F, Saus J, Antignac C. Splice-mediated insertion of an Alu sequence in the COL4A3 mRNA causing autosomal recessive Alport syndrome. Hum Mol Genet. 1995;4:675–679. doi: 10.1093/hmg/4.4.675. [DOI] [PubMed] [Google Scholar]
- 42.Koester J. Voltage-gated ion channels and generation of the action potential. In: Kandel E, Schwartz J, Jessell T, editors. Principles of neural science. New York, N.Y: Elsevier Science Publishing Co.; 1991. pp. 104–118. [Google Scholar]
- 43.Kohrman D C, Harris J B, Meisler M H. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J Biol Chem. 1996;271:17576–17581. doi: 10.1074/jbc.271.29.17576. [DOI] [PubMed] [Google Scholar]
- 44.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 45.Kolossova I, Padgett R A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA. 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 46.Krawczak M, Reiss J, Cooper D N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 47.Kreivi J P, Lamond A I. RNA splicing: unexpected spliceosome diversity. Curr Biol. 1996;6:802–805. doi: 10.1016/s0960-9822(02)00599-7. [DOI] [PubMed] [Google Scholar]
- 48.Kreivi J P, Zefrivitz K, Akusjärvi G. A U1 snRNA binding site improves the efficiency of in vitro pre-mRNA splicing. Nucleic Acids Res. 1991;19:6956. doi: 10.1093/nar/19.24.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo H C, Nasim F H, Grabowski P J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- 50.Lafrenière R G, Rochefort D L, Kibar Z, Fon E A, Han F, Cochius J, Kang X, Baird S, Korneluk R G, Andermann E, Rommens J M, Rouleau G A. Isolation and characterization of GT335, a novel human gene conserved in Escherichia coli and mapping to 21q22.3. Genomics. 1996;38:264–272. doi: 10.1006/geno.1996.0627. [DOI] [PubMed] [Google Scholar]
- 51.Lévy G, Levi-Acobas F, Blanchard S, Gerber S, Larget-Piet D, Chenal V, Liu X Z, Newton V, Steel K P, Brown S D, Munnich A, Kaplan J, Petit C, Weil D. Myosin VIIA gene: heterogeneity of the mutations responsible for Usher syndrome type IB. Hum Mol Genet. 1997;6:111–116. doi: 10.1093/hmg/6.1.111. [DOI] [PubMed] [Google Scholar]
- 52.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 53.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludwig D L, Mudgett J S, Park M S, Perez-Castro A V, MacInnes M A. Molecular cloning and structural analysis of the functional mouse genomic XPG gene. Mamm Genome. 1996;7:644–649. doi: 10.1007/s003359900198. [DOI] [PubMed] [Google Scholar]
- 55.Luukkonen B G, Séraphin B. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makalowski W, Mitchell G A, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 57.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 58.Matera A G, Ward D C. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McClatchey A I, Lin C S, Wang J, Hoffman E P, Rojas C, Gusella J F. The genomic structure of the human skeletal muscle sodium channel gene. Hum Mol Genet. 1992;1:521–527. doi: 10.1093/hmg/1.7.521. [DOI] [PubMed] [Google Scholar]
- 60.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCullough A J, Schuler M A. Intronic and exonic sequences modulate 5′ splice site selection in plant nuclei. Nucleic Acids Res. 1997;25:1071–1077. doi: 10.1093/nar/25.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mighell A J, Markham A F, Robinson P A. Alu sequences. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell G A, Labuda D, Fontaine G, Saudubray J M, Bonnefont J P, Lyonnet S, Brody L C, Steel G, Obie C, Valle D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc Natl Acad Sci USA. 1991;88:815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montzka K A, Steitz J A. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci USA. 1988;85:8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mori Y, Shiwaku H, Fukushige S, Wakatsuki S, Sato M, Nukiwa T, Horii A. Alternative splicing of hMSH2 in normal human tissues. Hum Genet. 1997;99:590–595. doi: 10.1007/s004390050411. [DOI] [PubMed] [Google Scholar]
- 66.Mount S M. AT-AC introns: an ATtACk on dogma. Science. 1996;271:1690–1692. doi: 10.1126/science.271.5256.1690. [DOI] [PubMed] [Google Scholar]
- 67.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 68.Newman A J. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nilsen T W. A parallel spliceosome. Science. 1996;273:1813. doi: 10.1126/science.273.5283.1813. [DOI] [PubMed] [Google Scholar]
- 70.Nilsen T W. RNA-RNA interactions in nuclear pre-mRNA splicing. In: Simons R W, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 279–307. [Google Scholar]
- 71.Niwa M, Berget S M. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 72.Ohshima T, Nagle J W, Pant H C, Joshi J B, Kozak C A, Brady R O, Kulkarni A B. Molecular cloning and chromosomal mapping of the mouse cyclin-dependent kinase 5 gene. Genomics. 1995;28:585–588. doi: 10.1006/geno.1995.1194. [DOI] [PubMed] [Google Scholar]
- 73.Ophoff R A, Terwindt G M, Vergouwe M N, van Eijk R, Oefner P J, Hoffman S M, Lamerdin J E, Mohrenweiser H W, Bulman D E, Ferrari M, Haan J, Lindhout D, van Ommen G J, Hofker M H, Ferrari M D, Frants R R. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- 74.Parker R, Siliciano P G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 75.Phillips M S, Fujii J, Khanna V K, DeLeon S, Yokobata K, de Jong P J, MacLennan D H. The structural organization of the human skeletal muscle ryanodine receptor (RYR1) gene. Genomics. 1996;34:24–41. doi: 10.1006/geno.1996.0238. [DOI] [PubMed] [Google Scholar]
- 76.Plant K E, Hair A, Morgan G T. Genes encoding isoforms of transcription elongation factor TFIIS in Xenopus and the use of multiple unusual RNA processing signals. Nucleic Acids Res. 1996;24:3514–3521. doi: 10.1093/nar/24.18.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plummer N W, Galt J, Jones J M, Burgess D L, Sprunger L K, Kohrman D C, Meisler M H. Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics. 1998;54:287–296. doi: 10.1006/geno.1998.5550. [DOI] [PubMed] [Google Scholar]
- 78.Rain J C, Rafi Z, Rhani Z, Legrain P, Krämer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richards A, Luccarini C, Pope F M. The structural organisation of LAMA4, the gene encoding laminin alpha4. Eur J Biochem. 1997;248:15–23. doi: 10.1111/j.1432-1033.1997.t01-1-00015.x. [DOI] [PubMed] [Google Scholar]
- 80.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Royaux I, Lambert de Rouvroit C, D’Arcangelo G, Demirov D, Goffinet A M. Genomic organization of the mouse reelin gene. Genomics. 1997;46:240–250. doi: 10.1006/geno.1997.4983. [DOI] [PubMed] [Google Scholar]
- 82.Ryan D G, Lin T, Brownie E, Bridger W A, Wolodko W T. Mutually exclusive splicing generates two distinct isoforms of pig heart succinyl-CoA synthetase. J Biol Chem. 1997;272:21151–21159. doi: 10.1074/jbc.272.34.21151. [DOI] [PubMed] [Google Scholar]
- 83.Sahba S, Nechiporuk A, Figueroa K P, Nechiporuk T, Pulst S M. Genomic structure of the human gene for spinocerebellar ataxia type 2 (SCA2) on chromosome 12q24.1. Genomics. 1998;47:359–364. doi: 10.1006/geno.1997.5131. [DOI] [PubMed] [Google Scholar]
- 84.Santisteban I, Arredondo-Vega F X, Kelly S, Debre M, Fischer A, Perignon J L, Hilman B, elDahr J, Dreyfus D H, Gelfand E W, et al. Four new adenosine deaminase mutations, altering a zinc-binding histidine, two conserved alanines, and a 5′ splice site. Hum Mutat. 1995;5:243–250. doi: 10.1002/humu.1380050309. [DOI] [PubMed] [Google Scholar]
- 85.Scadden A D J, Smith C W. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharp P A, Burge C B. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 87.Soldatov N M. Genomic structure of human L-type Ca2+ channel. Genomics. 1994;22:77–87. doi: 10.1006/geno.1994.1347. [DOI] [PubMed] [Google Scholar]
- 88.Souslova V A, Fox M, Wood J N, Akopian A N. Cloning and characterization of a mouse sensory neuron tetrodotoxin-resistant voltage-gated sodium channel gene, Scn10a. Genomics. 1997;41:201–209. doi: 10.1006/geno.1997.4669. [DOI] [PubMed] [Google Scholar]
- 89.Spector D L. Nuclear organization of pre-mRNA processing. Curr Opin Cell Biol. 1993;5:442–447. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- 90.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stamm S, Zhang M Q, Marr T G, Helfman D M. A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res. 1994;22:1515–1526. doi: 10.1093/nar/22.9.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stark J M, Bazett-Jones D P, Herfort M, Roth M B. SR proteins are sufficient for exon bridging across an intron. Proc Natl Acad Sci USA. 1998;95:2163–2168. doi: 10.1073/pnas.95.5.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stephens R M, Schneider T D. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J Mol Biol. 1992;228:1124–1136. doi: 10.1016/0022-2836(92)90320-j. [DOI] [PubMed] [Google Scholar]
- 94.Sterner D A, Carlo T, Berget S M. Architectural limits on split genes. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suchi M, Sano H, Mizuno H, Wada Y. Molecular cloning and structural characterization of the human histidase gene (HAL) Genomics. 1995;29:98–104. doi: 10.1006/geno.1995.1219. [DOI] [PubMed] [Google Scholar]
- 96.Takahara K, Lee S, Wood S, Greenspan D S. Structural organization and genetic localization of the human bone morphogenetic protein 1/mammalian tolloid gene. Genomics. 1995;29:9–15. doi: 10.1006/geno.1995.1209. [DOI] [PubMed] [Google Scholar]
- 97.Talerico M, Berget S M. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tarn W Y. Site-specific substitution of inosine at the terminal positions of a pre-mRNA intron: implications for the configuration of the terminal base interaction. Biochimie. 1996;78:1057–1065. doi: 10.1016/s0300-9084(97)86730-2. [DOI] [PubMed] [Google Scholar]
- 100.Tarn W Y, Steitz J A. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 101.Tarn W Y, Steitz J A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 102.Tarn W Y, Steitz J A. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22:132–137. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 103.Tarn W Y, Yario T A, Steitz J A. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
- 104.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 106.Treisman R, Orkin S H, Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- 107.Tumer Z, Vural B, Tonnesen T, Chelly J, Monaco A P, Horn N. Characterization of the exon structure of the Menkes disease gene using vectorette PCR. Genomics. 1995;26:437–442. doi: 10.1016/0888-7543(95)80160-n. [DOI] [PubMed] [Google Scholar]
- 108.Valcárcel J, Singh R, Green M R. Mechanism of regulated pre-mRNA splicing. In: Lamond A I, editor. Pre-mRNA processing. Heidelberg, Germany: Springer-Verlag; 1995. pp. 97–112. [Google Scholar]
- 109.Wagener R, Kobbe B, Paulsson M. Genomic organisation, alternative splicing and primary structure of human matrilin-4. FEBS Lett. 1998;438:165–170. doi: 10.1016/s0014-5793(98)01293-9. [DOI] [PubMed] [Google Scholar]
- 110.Wang Q, Li Z, Shen J, Keating M T. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- 111.Wassarman K M, Steitz J A. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol Cell Biol. 1992;12:1276–1285. doi: 10.1128/mcb.12.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 113.Watanabe A, Ikejima M, Suzuki N, Shimada T. Genomic organization and expression of the human MSH3 gene. Genomics. 1996;31:311–318. doi: 10.1006/geno.1996.0053. [DOI] [PubMed] [Google Scholar]
- 114.Winand N J, Panzer J A, Kolodner R D. Cloning and characterization of the human and Caenorhabditis elegans homologs of the Saccharomyces cerevisiae MSH5 gene. Genomics. 1998;53:69–80. doi: 10.1006/geno.1998.5447. [DOI] [PubMed] [Google Scholar]
- 115.Wu H J, Gaubier-Comella P, Delseny M, Grellet F, Van Montagu M, Rouzé R. Non-canonical introns are at least 10 (9) years old. Nat Genet. 1996;14:383–384. doi: 10.1038/ng1296-383. [DOI] [PubMed] [Google Scholar]
- 116.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 117.Wu Q. Mechanisms of AT-AC pre-mRNA splicing in vitro. Ph.D. thesis. Stony Brook: State University of New York at Stony Brook; 1998. [Google Scholar]
- 118.Wu, Q., and A. R. Krainer. Unpublished observations.
- 119.Wu Q, Krainer A R. U1-mediated exon definition interactions between AT-AC and GT-AG introns. Science. 1996;274:1005–1008. doi: 10.1126/science.274.5289.1005. [DOI] [PubMed] [Google Scholar]
- 120.Wu Q, Krainer A R. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA. 1997;3:586–601. [PMC free article] [PubMed] [Google Scholar]
- 121.Wu Q, Krainer A R. Purine-rich enhancers function in the AT-AC pre-mRNA splicing pathway and do so independently of intact U1 snRNP. RNA. 1998;4:1664–1673. doi: 10.1017/s1355838298981432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xue J, Jorgensen M, Pihlgren U, Rask L. The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol Biol. 1995;27:911–922. doi: 10.1007/BF00037019. [DOI] [PubMed] [Google Scholar]
- 123.Xue J, Rask L. The unusual 5′ splicing border GC is used in myrosinase genes of the Brassicaceae. Plant Mol Biol. 1995;29:167–171. doi: 10.1007/BF00019128. [DOI] [PubMed] [Google Scholar]
- 124.Yamada Y, Masuda K, Li Q, Ihara Y, Kubota A, Miura T, Nakamura K, Fujii Y, Seino S, Seino Y. The structures of the human calcium channel alpha 1 subunit (CACNL1A2) and beta subunit (CACNLB3) genes. Genomics. 1995;27:312–319. doi: 10.1006/geno.1995.1048. [DOI] [PubMed] [Google Scholar]
- 125.Yu Y T, Steitz J A. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc Natl Acad Sci USA. 1997;94:6030–6035. doi: 10.1073/pnas.94.12.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu Y T, Tarn W Y, Yario T A, Steitz J A. More Sm snRNAs from vertebrate cells. Exp Cell Res. 1996;229:276–281. doi: 10.1006/excr.1996.0372. [DOI] [PubMed] [Google Scholar]
- 127.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang M Q. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 129.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 130.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]