Culture-Independent Identification of Microorganisms That Respond to Specified Stimuli (original) (raw)
Abstract
A new approach that permits culture-independent identification of microorganisms that respond to specified stimuli was developed. This approach was illustrated by examination of microorganisms that grew in response to various nutrient supplements added to soil. A thymidine nucleotide analog, bromodeoxyuridine (BrdU), and supplements were added to soil and incubated for 3 days. DNA was extracted from the soil, and the newly synthesized DNA was isolated by immunocapture of the BrdU-labeled DNA. The unique perspective this approach offers was demonstrated by comparing the microbial community structures obtained from total soil DNA and the BrdU-labeled fraction in an rRNA gene (rDNA) analysis. The traditional total DNA analysis revealed no notable differences between the treatments, whereas the BrdU-labeled DNA showed significantly different banding patterns between the nutrient supplement treatments and compared with total DNA banding patterns. PCR primers were developed to specifically amplify the intergenic region of an rDNA sequence unique to the BrdU analysis of a phosphate supplement treatment. Amplification of DNA from all treatments using these primers showed that it was unique to the phosphate treatment and that it was present in both the total DNA and BrdU-labeled DNA fractions. This result demonstrates the promise of this new strategy, because it was able to permit identification of a sequence from a phosphate-responsive organism that was not discernable in the traditional total DNA community structure analysis.
The majority of extant microorganisms have yet to be described, and fewer than one percent of these can be cultured by standard methods (1, 7). The extent of this diversity was exemplified by a study which used genetic complexity measurements to calculate that 4,000 completely different bacteria genomes inhabited a Norwegian forest soil sample (19). Recently, descriptions of rRNA genes (rDNA) obtained from environmental samples have provided both a glimpse of this extensive microbial diversity and a tool to identify uncultured organisms (1, 14). However, progress beyond the classification stage of microbial ecology to the physiological stage has been primarily limited to community-level analyses such as microbial respiration, substrate utilization, N2 fixation, signature lipid biomarkers, and enzyme activities (9).
To correlate the activity of specific microorganisms with defined environmental or physiological parameters, Stahl et al. pioneered the use of rRNA hybridization probes to monitor population levels and shifts (17). However, this strategy requires the prior identification and characterization of meaningful sequences. Given the immense complexity of most natural microbial communities, identification of such sequences is a daunting task. A random analysis of rRNA or rDNA sequences from an environmental sample would likely lead to the identification of the dominant organisms in that community but not necessarily the ones involved in a particular physiological response. This report describes a new approach to bridge the gap between function and phylogeny by permitting the identification of populations that grow in response to specified or measured stimuli.
MATERIALS AND METHODS
Nutrient supplementation and DNA extraction.
One-gram soil samples were amended with four different nutrient supplement treatments. Supplements were added to sieved soil samples throughout a 3-day period and incubated in petri dishes at room temperature. On day 1, H2O and glucose were mixed into the soil samples (500 μl of H2O was added to treatments C and D, 550 μl of H2O was added to treatment B, and 50 μl of 888 mM glucose was added to treatments C and D). On day 2, H2O and supplements were mixed into the soil samples (250 μl of H2O was added to treatments B and C, 200 μl of H2O was added to treatment D, 50 μl of 100 mM bromodeoxyuridine [BrdU] was added to treatments A to D, and 50 μl of 89 mM KH2PO4 was added to treatment D). On day 3, H2O and supplements were mixed into the soil samples (50 μl of H2O was added to treatments B to D, and 50 μl of 100 mM BrdU was added to treatments A to D). On day 4, DNA was extracted from the soil samples with the FastDNA Kit For Soil as described by the manufacturer (BIO 101, Vista, Calif.) (4).
BrdU immunocapture.
Immunocapture of BrdU-labeled DNA was performed by a modification of the method described by Haider et al. (8). Twenty-five microliters of Dynabeads (M-450) coated with sheep anti-mouse immunoglobulin G was washed three times with 200 μl of phosphate-buffered saline–bovine serum albumin (PBS-BSA [PBS containing 0.1% BSA]) as described by the manufacturer (Dynal, Lake Success, N.Y.). These beads were resuspended in 84 μl of PBS-BSA-herring (PBS-BSA containing 5 mg of herring sperm DNA [Promega, Madison, Wis.] per ml) and incubated with 16 μl of anti-BrdU antibody (Boehringer Mannheim, Indianapolis, Ind.) for 60 min at room temperature while being rotated. This mixture was washed three times with 200 μl of PBS-BSA and resuspended in 80 μl of PBS-BSA-herring. Twenty microliters of denatured soil DNA (95°C for 5 min, 5 min on ice) was added to this suspension and incubated at room temperature for 120 min while being rotated. This mixture was washed three times with 200 μl of PBS-BSA and resuspended in 40 μl of H2O. This suspension was heated at 95°C for 5 min and cooled on ice for 5 min, and the non-bead fraction was collected by magnetic capture. After this work was completed, an alternative procedure to isolate BrdU-labeled DNA was described (20). Comparison of the two procedures showed that the protocol described by Urbach et al. (20) is preferable, as it isolated less unlabeled DNA (data not shown).
Community structure analysis.
Community structure analysis was performed by resolving PCR-amplified 16S-23S rRNA intergenic fragments on a denaturing polyacrylamide gel (5). The intergenic spacer region between the small- and large-subunit rRNA genes was amplified in 150-μl PCR mixtures with the following final concentrations or total amounts: 20 ng of soil DNA or 1 ng of BrdU-labeled DNA, 50 mM Tris (pH 8.3), 500 μg of BSA per ml, 2.5 mM MgCl2, 250 μM deoxynucleoside triphosphates (dNTPs), 200 nM forward primer 1406F TGYACACACCGCCCGT (10), 200 nM reverse primer 23R GGGTTBCCCCATTCRG, and 7.5 U of Taq DNA polymerase. All reagents were combined and heated at 94°C for 2 min. For the soil DNA, 25 cycles of PCR were performed; for the BrdU-labeled DNA, 35 cycles were performed. PCRs were done with an Air Thermo-Cycler (Idaho Technologies, Idaho Falls, Idaho) at 94°C for 15 s, 52°C for 15 s, and 72°C for 30 s, followed by 72°C for 2 min. PCR products were purified with the QIAquick PCR Purification Kit (Qiagen, Valencia, Calif.), eluted with 50 μl of H2O, dried to a volume of 10 μl, and denatured by addition of 10 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromphenol blue, 0.05% xylene cyanol FF) and heating at 95°C for 5 min. The DNA was loaded on a prerun 5% (29:1) polyacrylamide–0.6× Tris-borate-EDTA TBE gel (0.375 mm thick) containing 8 M urea and electrophoresed at 50 W for 150 min to maintain a temperature between 55°C and 60°C. Gels were silver stained with the SILVER SEQUENCE DNA Staining Reagents (Promega) (2).
Sequence-specific PCR.
Band A was excised from the polyacrylamide gel (see Fig. 2), PCR amplified with the parameters described above, cloned into the pGEM-T vector as described by the manufacturer (Promega), and sequenced with an ABI PRISM Dye Terminator Cycle Sequencing Kit (Perkin Elmer, Foster City, Calif.). Sequence-specific PCR primers were designed to hybridize to the 16S-23S rRNA intergenic region of this sequence. Ten-microliter PCRs were performed on total DNA and BrdU-labeled DNA from all treatments, resolved on a 1.25% agarose gel, and stained with ethidium bromide. PCRs contained 50 mM Tris (pH 8.3), 2.5 mM MgCl2, 500 ng of BSA per μl, 250 nM dNTPs, 400 nM forward primer (TCACTTACTGTTCGGTTT), 400 nM reverse primer (CTTGGATAGAAGAAGCAT), 0.5 U of Taq DNA polymerase, and either 25 ng of total DNA template or 1 ng of BrdU-captured DNA template. PCR cycle parameters were as follows: 2 min at 94°C, 40 cycles of 0 s at 94°C, 20 s at 50°C, and 10 s at 72°C; and 2 min at 72°C.
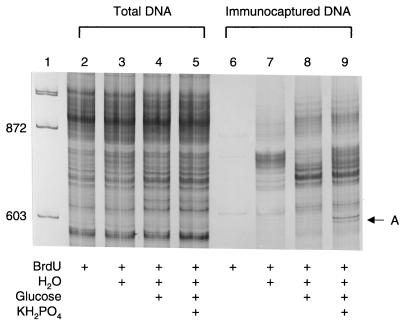
FIG. 2.
Comparison of the microbial community structures obtained from total DNA and BrdU-labeled DNA by analysis of the 16S-23S rRNA intergenic region after exposure to nutrient supplements. Lanes: 1, φX174/_Hae_III molecular weight marker (Promega); 2 and 6, treatment A; 3 and 7, treatment B; 4 and 8, treatment C; 5 and 9, treatment D.
Nucleotide sequence accession number.
The 16S-23S rRNA intergenic sequence obtained from band A (see Fig. 2) has been deposited in GenBank under the accession number AF124217.
RESULTS AND DISCUSSION
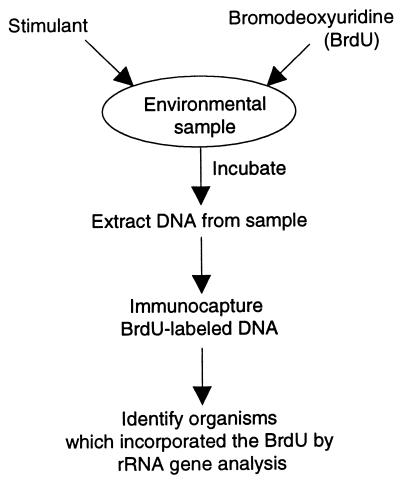
This recently designed methodology uses a modification of the thymidine incorporation technique, which quantifies microbial growth by measuring the amount of thymidine that is incorporated into microbial DNA (13). [3H]thymidine uptake has been used successfully to measure in situ bacterial growth in soil, aquatic, and other environments (6, 13, 18). By replacing [3H]thymidine with BrdU, the newly synthesized DNA can be isolated by BrdU immunocapture. The microbes that grow in response to a stimulant may therefore be identified by (i) the simultaneous addition of BrdU and a stimulant to an environmental sample, (ii) sample incubation, (iii) DNA extraction, (iv) immunocapture of the BrdU-labeled DNA, and (v) rRNA gene analysis (Fig. 1).
FIG. 1.
Outline of the strategy to identify microorganisms that respond to stimuli.
To depict the unique perspective that can be obtained from this approach, the microbial community structures of soil supplemented with four different nutrients were examined by both the new BrdU-labeled DNA strategy and a traditional total DNA analysis (Fig. 2). The community analyses were done by resolving PCR-amplified 16S-23S rRNA intergenic region fragments on a denaturing polyacrylamide gel (5). The size heterogeneity of these fragments provides a simple method to depict bacterial community structure. When comparing the banding patterns from the four treatments by use of total DNA, virtually no difference can be seen. However, the BrdU-labeled DNA revealed significantly different patterns, both between the treatments and compared to that of the total DNA. A likely explanation for the difference is that the standard total DNA analysis examines all of the DNA extracted from soil, including DNA from organisms that did not respond to the treatment and naked DNA that is associated with soil particles. Conversely, the BrdU-labeled DNA approach examines only newly synthesized DNA, allowing the identification of the responding and actively growing organisms. In addition, the lack of PCR products in the BrdU-only (lane 6) and no-BrdU (data not shown) treatments shows that the BrdU did not act as a stimulant for growth and that the immunocapture process is specific.
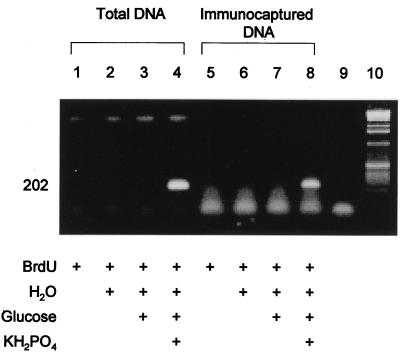
To validate these different depictions of bacterial diversity, the relative abundance of a specific sequence unique to the phosphate treatment was assessed. This sequence was obtained by cloning the 16S-23S rRNA intergenic region DNA contained in band A (Fig. 2, lane 9). The clone used in this analysis has 96% sequence similarity to a 16S rRNA gene from Bacillus subtilis (12). To verify its relative abundance, specific PCR primers were designed to hybridize to the 16S-23S rRNA intergenic region of band A. Both total DNA and BrdU-captured DNA from all treatments were subjected to amplification by PCR (Fig. 3). Only the phosphate treatment produced a PCR product, suggesting that the organism represented by band A specifically responded to the phosphate supplement. The fact that band A was successfully amplified in both the BrdU-captured DNA and the total DNA demonstrates the promise of this new strategy, as it identified a sequence from a phosphate-responsive organism that was not discernable in the traditional total DNA analysis. The significant genetic complexity of total DNA analyses will presumably lead to depictions of microbial community structure biased towards the most numerous gene sequences, which may have no relevance to a given physiological response. The BrdU capture approach removes large pools of background DNA, enabling analysis of only actively growing populations.
FIG. 3.
Validation of a specific rRNA gene sequence from a phosphate-responsive organism by PCR. Lanes: 9, no-template negative control; 10, 1-kb ladder (Gibco BRL, Grand Island, N.Y.).
Recently, Urbach et al. (20) demonstrated the promise of the BrdU strategy in an aquatic environment. They showed that the BrdU approach isolated a subset of the total DNA located in a freshwater lake, suggesting that only a fraction of this bacterial community was actively dividing. My study expands on this concept by demonstrating the utility of this strategy to examine the effects of exogenous chemical stimuli on bacterial community structure in soil. I also have shown the advantage of the BrdU strategy over a total rDNA analysis in identifying changes in community structure. The potential limitations of the BrdU strategy described in both reports include the range of organisms capable of BrdU uptake and incorporation. Urbach et al. (20) showed that only two of four different bacterial strains incorporated BrdU. Other reports, however, suggest that the majority of bacteria take up and incorporate [3H]thymidine (16). Since BrdU has been used successfully as a thymidine analog in numerous applications, it is likely that most organisms will also take up and incorporate BrdU. Another possible limitation of this strategy may come from the potential toxicity of halogenated nucleotides incorporated into DNA (11). However, the growth kinetics for specific bacterial cultures obtained with and without BrdU were shown to be similar (20). In addition, if used at modest concentrations, BrdU appears to be relatively safe and is currently being administered to human cancer patients (3, 15). Despite these potential shortcomings, both of these reports demonstrate an approach that can assist in the investigation of uncultured microorganisms. In future work, this strategy could be modified to include the identification of important genes, since the genomic DNAs of the responding microorganisms are also captured. It could also be used to identify microorganisms that respond to measurable natural phenomena such as nutrient availability and signal molecules.
ACKNOWLEDGMENTS
I thank Gary G. Judd and Donald A. Cooksey for their comments on the manuscript.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 3.Begg A C, McNally N J, Shrieve D C, Karcher H. A method to measure the duration of DNA synthesis and the potential doubling time from a single sample. Cytometry. 1985;6:620–626. doi: 10.1002/cyto.990060618. [DOI] [PubMed] [Google Scholar]
- 4.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brock T D. Bacterial growth rate in the sea: direct analysis by thymidine autoradiography. Science. 1967;155:81–83. doi: 10.1126/science.155.3758.81. [DOI] [PubMed] [Google Scholar]
- 7.Colwell R R, Clayton R A, Ortiz-Conde B A, Jacobs D, Russek-Cohen E. The microbial species concept and biodiversity. In: Allsopp D, Colwell R R, Hawksworth D L, editors. Microbial diversity and ecosystem function. Oxon, United Kingdom: CAB International; 1995. pp. 3–15. [Google Scholar]
- 8.Haider S R, Juan G, Traganos F, Darzynkiewicz Z. Immunoseparation and immunodetection of nucleic acids labeled with halogenated nucleotides. Exp Cell Res. 1997;234:498–506. doi: 10.1006/excr.1997.3644. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy A C, Papendick R I. Microbial characteristics of soil quality. J Soil Water Conserv. 1995;50:243–248. [Google Scholar]
- 10.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: Wiley; 1991. pp. 115–175. [Google Scholar]
- 11.Li X, Patel R, Melamed M R, Darzynkiewicz Z. The cell cycle effects and induction of apoptosis by 5-bromouridine in cultures of human leukaemic molt-4 and HL-60 cell lines and mitogen-stimulated normal lymphocytes. Cell Prolif. 1994;27:307–319. doi: 10.1111/j.1365-2184.1994.tb01428.x. [DOI] [PubMed] [Google Scholar]
- 12.Loughney K, Lund E, Dahlberg J E. tRNA genes are found between the 16s and 23s rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982;10:1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriarty D J W. Measurement of bacterial growth rates in aquatic systems from rates of nucleic acid synthesis. Adv Microb Ecol. 1986;9:246–292. [Google Scholar]
- 14.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 15.Raza A G, Miller M, Mazewski C, Sheikh Y, Lampkin B, Sawaya R, Crone K, Berger T, Reisling J, Gray J, Khan S, Preisler H D. Observations regarding DNA replication sites in human cells in vivo following infusions of iododeoxyuridine and bromodeoxyuridine. Cell Prolif. 1991;24:113–126. doi: 10.1111/j.1365-2184.1991.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 16.Robarts R D, Zohary T. Fact or fiction—bacterial growth rates and production as determined by (methyl-3H)-thymidine? Adv Microb Ecol. 1993;13:371–425. [Google Scholar]
- 17.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas D R, Richardson J A, Dicker R J. The incorporation of tritiated thymidine into DNA as a measure of the activity of soil micro-organisms. Soil Biol Biochem. 1974;6:293–296. [Google Scholar]
- 19.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbach E, Vergin K L, Giovannoni S J. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl Environ Microbiol. 1999;65:1207–1213. doi: 10.1128/aem.65.3.1207-1213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]