Lysine 2,3-Aminomutase from Clostridium subterminale SB4: Mass Spectral Characterization of Cyanogen Bromide-Treated Peptides and Cloning, Sequencing, and Expression of the Gene kamA in Escherichia coli (original) (raw)
Abstract
Lysine 2,3-aminomutase (KAM, EC 5.4.3.2.) catalyzes the interconversion of l-lysine and l-β-lysine, the first step in lysine degradation in Clostridium subterminale SB4. KAM requires _S_-adenosylmethionine (SAM), which mediates hydrogen transfer in a mechanism analogous to adenosylcobalamin-dependent reactions. KAM also contains an iron-sulfur cluster and requires pyridoxal 5′-phosphate (PLP) for activity. In the present work, we report the cloning and nucleotide sequencing of the gene kamA for C. subterminale SB4 KAM and conditions for its expression in Escherichia coli. The cyanogen bromide peptides were isolated and characterized by mass spectral analysis and, for selected peptides, amino acid and N-terminal amino acid sequence analysis. PCR was performed with degenerate oligonucleotide primers and C. subterminale SB4 chromosomal DNA to produce a portion of kamA containing 1,029 base pairs of the gene. The complete gene was obtained from a genomic library of C. subterminale SB4 chromosomal DNA by use of DNA probe analysis based on the 1,029-base pair fragment. The full-length gene consisted of 1,251 base pairs specifying a protein of 47,030 Da, in reasonable agreement with 47,173 Da obtained by electrospray mass spectrometry of the purified enzyme. N- and C-terminal amino acid analysis of KAM and its cyanogen bromide peptides firmly correlated its amino acid sequence with the nucleotide sequence of kamA. A survey of bacterial genome databases identified seven homologs with 31 to 72% sequence identity to KAM, none of which were known enzymes. An E. coli expression system consisting of pET 23a(+) plus kamA yielded unsatisfactory expression and bacterial growth. Codon usage in kamA includes the use of AGA for all 29 arginine residues. AGA is rarely used in E. coli, and arginine clusters at positions 4 and 5, 25 and 27, and 134, 135, and 136 apparently compound the barrier to expression. Coexpression of E. coli argU dramatically enhanced both cell growth and expression of KAM. Purified recombinant KAM is equivalent to that purified from C. subterminale SB4.
KAM from Clostridium subterminale SB4 catalyzes the interconversion of l-lysine and l-β-lysine. In C. subterminale SB4, the production of β-lysine is the first step in the metabolism of lysine as the sole source of carbon and nitrogen, leading to acetate and butyrate as final products (39). Unlike other aminomutases, including d-lysine 5,6-aminomutase (1, 26), d-ornithine 2,3-aminomutase (2), and l-leucine 2,3-aminomutase (33), KAM is not adenosylcobalamin dependent. Instead, the enzyme is activated by SAM and contains iron-sulfur clusters and PLP (7, 31, 38).
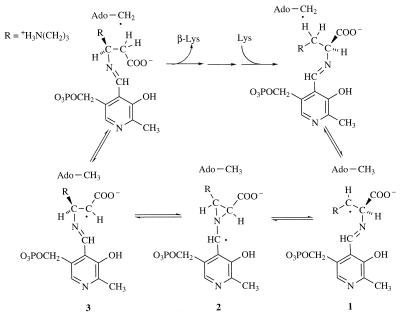
The interconversion of l-α-lysine and l-β-lysine is a reversible process in which an unactivated, carbon-bound hydrogen atom undergoes a 1,2-migration concomitant with the countermigration of the α-amino group by a novel free-radical-based mechanism (12, 13). According to the current working hypothesis, homolytic cleavage of the _S_-adenosyl bond in SAM is mediated by the [4Fe-4S] center and produces the 5′-deoxyadenosyl radical as a transient intermediate (27, 28). As illustrated in Fig. 1, the 5′-deoxyadenosyl radical initiates the rearrangement by abstracting the 3-pro-R hydrogen of l-lysine in the forward direction to form radical 1 or the 2-pro-R hydrogen of l-β-lysine in the reverse direction to form radical 3. Evidence suggests that both amino acids are bound to the enzyme in the form of external aldimines with PLP. The substrate- and product-related free radicals are interconverted by way of an azacyclopropylcarbinyl, radical intermediate 2.
FIG. 1.
The mechanism of the radical rearrangement catalyzed by KAM.
KAM was first observed by Costilow and coworkers in crude extracts of C. subterminale SB4 (8) and later was purified and found to contain PLP and iron and to require SAM for activity (7). Enzyme activity was rapidly destroyed by dioxygen. The purified enzyme was partially inactive and could be further activated by prolonged anaerobic incubation with PLP, iron, and glutathione or dihydrolipoate, followed by addition of SAM and dithionite (7). KAM is a multisubunit protein with a subunit _M_r of 48,000 and an overall _M_r of 285,000 (7, 38). Later studies demonstrated the presence of labile sulfide in the form of three [4Fe-4S] centers (31) and six molecules of PLP per hexamer (38).
Although KAM has been described as a hexamer of identical subunits, the amino acid composition and the precise molecular weight have not been established. Furthermore, the subunit composition has not been fully confirmed. Because the most highly purified samples displayed minor bands upon analysis by SDS-PAGE and the cofactor requirements were complex, the possibility that an additional subunit might be involved could not be ruled out. Although the principal band observed by gel electrophoresis seemed to correspond to a single protomer in a hexameric enzyme, the iron and sulfide content and electron paramagnetic resonance spectral analysis indicated just three [4Fe-4S] centers per hexamer, suggesting that the subunits might not be identical. In order to examine the molecular composition of KAM, the purified enzyme has been characterized by CNBr fragmentation and amino acid sequence and mass spectral analysis. The clostridial gene has been cloned and expressed in Escherichia coli. Expression was dramatically facilitated by coexpression of argU, which encoded and enhanced the production of the most rare species of tRNAArg in E. coli. The recombinant KAM proved to be indistinguishable from KAM purified from clostridia, confirming the subunit composition as a homohexamer.
MATERIALS AND METHODS
Abbreviations.
The abbreviations used are as follows: KAM, lysine 2,3-aminomutase; SAM, _S_-adenosylmethionine; PLP, pyridoxal 5′-phosphate; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; Epps, 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid; DTT, dithiothreitol; IPTG, isopropyl-β-d-thiogalactopyranoside; X-Gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; PITC, phenylisothiocyanate; GuHCl, guanidine hydrochloride; TFA, trifluoroacetic acid; CNBr, cyanogen bromide; HPLC, high-pressure liquid chromatography; LB, Luria-Bertani; ARR, anaerobic ribonucleotide reductase; PFL, pyruvate formate lyase.
Materials.
Tris, Epps, HEPES, EDTA, DTT, TFA, carbenicillin, PLP, dihydrolipoic acid, and SAM were purchased from Sigma Chemical Co. SAM was further purified as described by Lieder et al. (25). Tetracycline hydrochloride, HPLC-grade acetonitrile, and methanol were purchased from Fisher. CNBr and 4-vinylpyridine were purchased from Aldrich Chemical Co. IPTG and X-Gal were purchased from Promega, ampicillin was from Boehringer Mannheim, PITC was from Pierce Chemical Co., and GuHCl was from Life Technologies. l-[U-14C]lysine (55,000 dpm, >300 Ci/mol) was purchased from New England Nuclear. For sequencing, [α-35S]dATPαS and the T7 Sequenase version 2.0 DNA sequencing kit were purchased from Amersham Life Science. Phenyl Sepharose CL-6B and Q Sepharose Fast Flow were obtained from Amersham Pharmacia Biotech.
Enzymes, bacterial strains, and plasmids.
Restriction endonucleases were purchased as follows. _Nde_I, _Xho_I, _Bam_HI, and _Xba_I were from Promega and _Hin_dIII, _Eco_RI, and _Acc_I were from New England Biolabs. Taq Plus long DNA polymerase and Pfu DNA polymerase were purchased from Stratagene. T4 DNA ligase and calf intestine alkaline phosphatase were purchased from Amersham Pharmacia Biotech, and deoxyribonuclease I was from Sigma Chemical Co. Plasmid vectors were purchased as follows: pCR2.1 was from Invitrogen, pUC19 was from New England Biolabs, pET23a(+) was from Novagen, pAlter-Ex2 was from Promega, and pCR-Script SK(+) Amp was from Stratagene. Competent E. coli cells were purchased as follows: XL-2 Blue Ultracompetent, XL-1 Blue MRF′, XL-2 Blue MRF′, and JM109 were from Stratagene and BL21(DE3) was from Novagen.
Purification of KAM.
KAM from C. subterminale SB4 and recombinant KAM from E. coli were purified by the procedure of Moss and Frey (28), as modified by Petrovich et al. (32). KAM was frozen as small pellets in liquid N2 and stored in liquid N2.
Analytical methods.
The activity of KAM was measured by the procedure of Chirpich et al. (7) as amended (4). The following additional modifications were made. For reductive incubation, enzyme (1 μM) was incubated for 4 h at 37°C in 0.04 M Na Epps buffer (pH 8.0) with 1 mM ferric ammonium citrate, 0.5 mM PLP, and 20 μM dihydrolipoic acid in a total volume of 0.36 ml. For the activity assay, reductively incubated enzyme was diluted fivefold with 0.2 M Na Epps buffer (pH 8.0), and 25 μl was added to 30 μl of 40 mM l-[U-14C]lysine, 18 μM SAM, and 3 mM sodium dithionite in 0.3 M Na Epps buffer (pH 8.0) (0.09 μM was the final enzyme concentration). Enzyme samples were incubated at 37°C for 4 to 6 min. The reaction was quenched by the addition of 30 μl of 0.2 M formic acid. Paper electrophoresis and scintillation counting were done as described previously (4).
The concentration of purified KAM was determined spectrophotometrically, using the value ɛ280 = 3.6 × 105 M−1 cm−1 (38). Metal analyses were carried out by inductively coupled plasma emission mass spectrometry in the Plant and Soil Analysis Laboratory of the University of Wisconsin—Madison. N- and C-terminal amino acid sequence analysis was performed at the Department of Biochemistry Macromolecular Facility, Michigan State University, East Lansing. SDS-PAGE of purified proteins was run on 10 to 20% polyacrylamide gradient Tris-HCl Ready gels (Bio-Rad). Gel electrophoresis under nondenaturing conditions was run in the same buffer, with the exclusion of SDS.
CNBr cleavage of KAM.
Purified C. subterminale SB4 KAM (subunit concentration, 200 μM) was dialyzed overnight (1 volume of protein to 1,000 volumes of 1 mM NaCl) and lyophilized. The dried protein was resuspended to the original volume in 6 M GuHCl–0.25 M Tris-HCl (pH 8.5)–1 mM EDTA. The protein was reduced with DTT (fivefold molar excess of DTT over cysteine residues) for 3 h at 25°C under argon and alkylated with 4-vinylpyridine (20-fold molar excess over DTT) for 90 min at 25°C. The protein was dialyzed against distilled water (1 volume of protein to 1,000 volumes of water) overnight at 4°C and lyophilized. The dried protein was dissolved in 0.1 N hydrochloric acid and subjected to CNBr cleavage by the addition of 100-fold molar excess of CNBr relative to methionine residues under argon for 24 h at 25°C. The sample was dried by Speed-Vac (Savant Instruments) under vacuum conditions and redissolved in 6 M GuHCl.
Peptide mapping by HPLC.
HPLC of CNBr-treated peptides was conducted with a C4 reversed-phase column (Vydac 214TP54, 5 μm, 4.6 by 250 mm; The Separations Group). The polypeptides were first separated into five main groups with a linear gradient of 0 to 80% acetonitrile in 0.1% TFA in water over 60 min at a flow rate of 1 ml/min at room temperature. The groups were designated peptide 1 (27 min), peptide 2 (33 min), peptides 3A and 3B (47 min), peptides 4A, 4B, and 4C (52 min), and peptide 5 (58 min). Fractions were analyzed spectrophotometrically at 214 nm, and selected fractions were evaporated by Speed-Vac, reinjected into the same column, and eluted with the following linear gradients of acetonitrile in 0.1% TFA in water at 1 ml/min: peptide 1, 5 to 20% for 1 h; peptide 2, 5 to 25% for 1 h; peptides 3A and 3B, 30 to 42% for 6 h; peptide 4A, 33 to 50% for 6 h; peptides 4B and 4C, 33 to 42% for 6 h; and peptide 5, 45 to 55% for 6 h.
Polypeptides were analyzed for amino acids by acid (HCl) hydrolysis, derivatization of the amino acids by reaction with PITC, and separation and quantification of individual PITC-amino acids. Derivatization, separation, and quantification of amino acids were conducted according to methods by Heinrikson and Meredith (20).
Cloning and sequencing the gene for KAM from C. subterminale SB4.
N-terminal amino acid sequence information from KAM and one of its CNBr-treated peptides (peptide 3A) was used to design two degenerate oligonucleotides, which served as primers for PCR. The N-terminal sequence of KAM used for primer design was KDVSDA, corresponding to the DNA plus strand 5′-AARGAYGTIWSIGAYGC-3′, where I is inosine, S is G+C, W is A+T, Y is C+T, D is G+A+T, and R is A+G. The N-terminal sequence of peptide 3A used for primer design was QSHDKV, corresponding to the opposite minus strand 5′-ATIACYTTRTCRTGISWYTG-3′.
PCR was used to generate an oligonucleotide of 1,029 bases which, when cloned and sequenced, yielded approximately 82% of the gene. Chromosomal DNA from C. subterminale SB4 was prepared and purified with a genomic tip 550/G kit (Qiagen, Inc.). PCR was carried out with Stratagene Taq Plus polymerase with low-salt buffer provided and 2 μg of C. subterminale DNA. All samples were subjected to 35 cycles of 1 min at 94°C, 30 s at 37°C, 15 s at 50°C, and 3 min at 72°C. After thermocycling, DNA formed during the PCR process was purified by agarose electrophoresis (2% agarose) in 0.04 M Tris-acetate (pH 8.0) with 1 mM EDTA. DNA obtained from PCR was cloned directly into the pCR2.1 vector (TA cloning kit, Invitrogen) according to the manufacturer's procedure.
The PCR insert was sequenced in both strands beginning at the ligation sites by the radiolabeled dideoxynucleotide method of Sanger et al. (35), using a T7 Sequenase version 2.0 sequencing kit (Amersham Life Science). The remaining unknown sequence of the gene was obtained by preparing a genomic library of C. subterminale SB4 chromosomal DNA. A nondegenerate, nonradioactive probe (500 bases) containing digoxigenin dUMP residues randomly incorporated was prepared by PCR with the PCR DIG Probe synthesis kit (Boehringer Mannheim). The following primers were used for the PCR Probe synthesis kit: primer 1 plus strand, 5′-ATCCTAACGATCCTAATGATCC-3′, and primer 2 minus strand, 5′-TGGATGGTTAAAGTGAGTG-3′. Using as the template a plasmid containing an incomplete kamA gene, a probe labeled randomly with digoxigenin groups was prepared.
Ten micrograms of C. subterminale SB4 chromosomal DNA was digested with 100 U each of _Eco_RI, _Xba_I, _Acc_I, and _Nde_I in separate reaction mixtures for 90 min at 37°C in eight replicates. These enzymes did not cut in the region of the known kamA nucleotide sequence, but their recognition sites were present in the multicloning region of pUC19. After restriction digestion, each reaction product was applied to a preparative agarose gel and subjected to electrophoresis. Several lanes were separated from the remaining gel for probe analysis according to the manufacturer's (Boehringer Mannheim) procedure. Positive probe-template interaction was identified by chemiluminescence from an anti-digoxigenin antibody conjugate containing alkaline phosphatase and reacting with CDP-Star (disodium 2-chloro-5-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.1.]decan}-4-yl)-1-phenyl phosphate) obtained from Boehringer Mannheim. Probe-positive gel regions were excised from the remaining agarose gel to create subgenomic libraries. DNA was extracted from the agarose by use of spin columns (Genelute agarose spin column; Supelco) as directed and then was concentrated by ethanol precipitation.
Chromosomal DNA fragments were ligated into restriction endonuclease-cut, dephosphorylated, and gel-purified pUC19 plasmid vector, transformed into competent E. coli XL-2 Blue Ultracompetent cells, and plated onto LB agar–carbenicillin–X-Gal–IPTG.
The subgenomic library was prepared by ligating 10 ng of prepared vector with either a stoichiometric or threefold excess of purified insert DNA. E. coli XL-2 Blue Ultracompetent cells were transformed with each ligation mix, and white colonies were plated in replication. One set of colonies from each library was transferred to nylon membranes, treated with alkali, and hybridized with oligonucleotide probe labeled with digoxigenin dUMP. Colonies (1 or 2 per 500) exhibiting chemiluminescence were chosen for further screening by DNA sequencing. The start codon, ATG, was found in one _Xba_I colony (X158). The start (ATG) and stop (TAA) codons were found in one _Eco_RI colony (E138). Double-stranded DNA from these selected colonies was sequenced by the automated ABI Prism Dye terminator cycle sequencing procedure (University of Wisconsin Biotechnology Center, Madison) to obtain the final sequence of the C. subterminale SB4 KAM gene (kamA).
Sequence alignments were done with the PileUp computer program of Genetics Computer Group, Madison, Wis.
Preparation of an E. coli expression vector for the C. subterminale SB4 KAM gene.
kamA was inserted into pET23a(+) for expression. In order to splice kamA into the vector with the start codon correctly spaced from the ribosome binding site, PCR was used to generate an insert which, after appropriate restriction digestion, could be cloned directly into the multicloning site. The pUC19 vector, E138, which contained the nucleotide sequence of the entire gene from the genomic library, was used as template. The following primers for PCR with pET23a(+) were used: plus strand, 5′-TACACATATGATAAATAGAAGATATG-3′, and minus strand, 5′-TAGACTCGAGTTATTCTTGAACGTGTCTC-3′. The PCR mixture (total volume, 100 μl) contained the following: pUC19 plasmid DNA (E138), 400 ng; deoxynucleoside triphosphates, 0.2 mM concentrations of each; oligonucleotide primers, 1 μM concentrations of each; and cloned Pfu DNA polymerase, 5 U. All samples were subjected to 35 cycles of 1 min at 94°C, 30 s at 37°C, 15 s at 50°C, and 3 min at 72°C. After thermocycling, DNA formed during the PCR process was purified by agarose electrophoresis (2% agarose). The purified PCR product was blunt-end ligated to pCR-Script Amp cloning vector according to the manufacturer's specification.
Plasmid DNA was double digested with _Nde_I/_Xho_I, and the kamA fragment was gel purified as described before. pET-23a(+) (10 μg) was similarly digested with _Nde_I/_Xho_I, dephosphorylated with 1 U of calf intestine alkaline phosphatase for 30 min at 37°C, gel purified, and ethanol precipitated. kamA and cut pET-23a(+) were ligated with T4 DNA ligase. Plasmids from individual colonies were sequenced in their entirety, including both regions of the start and stop codons to confirm the correctness of the construct, which was named pAF-80/kamA. For expression of the C. subterminale SB4 kamA gene in E. coli, pAF-80/kamA was transformed into competent BL21(DE3) E. coli cells.
Preparation of an expression vector for the arginyl tRNA gene, argU.
The sequence of the E. coli argU gene was published by Garcia et al. (16). The argU gene was isolated from E. coli chromosomal DNA by PCR. Primers which produced a 327-bp insert containing _Bam_HI and _Eco_RI restriction sites necessary for cloning into the pAlter-Ex2 plasmid vector under control of the tac promoter were prepared. This vector has a p15a origin of replication which allows it to be maintained with colE1 vectors, such as pET-23a(+). Also, the presence of this vector confers tetracycline resistance to E. coli. The PCR primers used were 5′-TATAGGATCCGACCGTATAATTCACGCGATTACACC-3′ (plus strand) and 5′-TAGAGAATTCGATTCAGTCAGGCGTCCCATTATC-3′ (minus strand). Chromosomal DNA from E. coli JM109 was prepared and purified with the Qiagen genomic tip 500/G kit. The PCR mixture (total volume, 100 μl) contained the following: E. coli chromosomal DNA, 2.5 μg; cloned Pfu DNA polymerase reaction buffer (Stratagene); deoxynucleoside triphosphates, 0.2 mM concentrations of each; oligonucleotide primers, 1 μM concentrations of each; and cloned Pfu DNA polymerase, 5 U. Reactions were cycled, and the resulting ∼320-bp band was gel purified as described before. The purified PCR product was blunt-end ligated to the pCR-Script Amp cloning vector with 0.3 pmol of vector according to the manufacturer's specifications. Plasmid DNA from the construct was digested with _Bam_HI and _Eco_RI, and the insert DNA was gel purified. The expression vector pAlter-Ex2 was similarly cut with _Bam_HI and _Eco_RI and prepared as pUC19. The argU insert and the pAlter-Ex2 cut vector were ligated with T4 DNA ligase as before. Competent BL21(DE3) cells were transformed and plated onto LB agar–tetracycline (12.5 μg/ml). Plasmid DNA was isolated, and the insert was sequenced completely by the Sequenase method.
Preparation of an expression system containing kamA and argU vectors.
Cells of BL21(DE3)/pAlter-Ex2 argU were made competent as previously described (34). Competent BL21(DE3)/pAlter-Ex2 _argU_-containing cells with the pAlter-Ex2 vector (argU gene) were transformed with pAF-80/kamA and plated on LB agar–carbenicillin (100 μg/ml)–tetracycline (10 μg/ml) overnight at 37°C.
Cell culture for protein expression.
Individually plated colonies of E. coli cells containing the various expression vectors were grown aerobically in 5 ml of LB medium containing appropriate antibiotics with or without IPTG (1 mM) overnight at 37°C. Sonicated cells and cell extracts following centrifugation at 14,000 × g for 30 min at 4°C were analyzed by SDS-PAGE on 10 to 20% gradient polyacrylamide gels and stained with Coomassie blue R-250.
Anaerobic expression of recombinant KAM in E. coli.
For purification of recombinant KAM, E. coli cells containing the expression vector pAF-80/kamA, with or without pAlter-Ex2/argU, were grown anaerobically in a glass fermentor (VirTis Co.) containing 15 liters of 2× YT medium (16 g of Bacto Tryptone, 10 g of Bacto Yeast Extract, 5 g of NaCl [per liter]) supplemented with 50 μM Fe(II)SO4, 50 μM ZnSO4, 50 μM Na2S, 4 mM sodium mercaptoacetate, and ampicillin (100 μg/ml), with or without tetracycline (10 μg/ml). The sealed flask was made anaerobic by gentle bubbling of N2 for 3 h prior to inoculation (1 ml of aerobic overnight cell stock). After approximately 14 h at 37°C, d-glucose was added to make a 0.2% (wt/vol) concentration. The culture was allowed to continue growing until reaching an optical density at 600 nm of 0.6 to 0.7, and then 1 mM IPTG was added to induce further expression of kamA. After 4 h, the culture was cooled to 24°C and allowed to continue growing for an additional 12 h before cell harvesting by concentration with tangential flow filtration (Pellicon System; Millipore Corp.), followed by centrifugation at 5,000 × g for 20 min. Cell pellets were quickly frozen in liquid N2 and stored in liquid N2. Approximately 30 to 35 g of cells was harvested from each 15-liter fermentation.
Nucleotide sequence accession number.
The nucleotide sequence of C. subterminale kamA has been deposited in GenBank, accession no. AF159146.
RESULTS
CNBr cleavage and purification of peptides.
In preparation for CNBr cleavage, the N-terminal amino acid sequence of purified KAM was obtained and found to be MINRRYELFKDVSDAD and the C-terminal sequence was found to be EQV. The finding of single N- and C-terminal sequences confirmed that the protein consisted of identical subunits. CNBr-treated peptides were prepared because the amino acid analysis indicated the presence of eight methionine residues, based on the assumption of identical subunits (38), of which all but one would contain homoserine lactone at their C termini (10). The limited number of CNBr-cleaved peptides facilitated the purification of an internal peptide for N-terminal sequence analysis.
Eight peptides produced by CNBr cleavage of clostridial KAM were separated by HPLC with a Vydac C4 reversed-phase column and several gradient elutions. The peptides were first separated into five main groups by a linear gradient of 0 to 80% acetonitrile in 0.1% TFA in water. Two of the eight peptides were homogeneous after this elution (peptides 1 and 2). The other peptides required rechromatography with narrower elution gradients. Five more homogeneous peptides were obtained.
One additional peptide fraction (peptide 3A) could not be resolved as a single peak on the chromatogram. Extensive attempts at separating this fraction into individual peptides with more shallow gradients were not successful. Subsequently, amino acid analysis of peptide 3A fractions collected at the early, middle, and late elution phases yielded essentially identical amino acid analyses (data not shown), suggesting that the peptides were essentially identical, perhaps differing in conformation. Analysis by electrospray mass spectrometry demonstrated a single mass for this peptide (Table 1), confirming that the multiple peaks arose from chromatographic rather than compositional heterogeneity. Peptide 3A was submitted for N-terminal amino acid sequence analysis and produced the single sequence PNYVISQSHDKV, confirming homogeneity and providing an internal sequence for use in cloning the gene.
TABLE 1.
Characteristics of the subunit and CNBr-cleaved and -alkylated peptides of KAM
| Peptide | Position | Mol wt (Da) | pIc | |
|---|---|---|---|---|
| Measureda | Calculatedb | |||
| N-terminal Metd | 1 | |||
| 4C (N terminal) | 2–57 | 6,972 | 6,972 | 5.4 |
| 4B | 58–124 | 7,403 | 7,403 | 4.3 |
| 6 | 125–127 | |||
| 1 | 128–145 | 2,352 | 2,352 | 9.0 |
| 7 | 146–147 | |||
| 5 | 148–218 | 8,002 | 8,002 | 5.2 |
| 3B | 219–272 | 6,229 | 6,230 | 7.1 |
| 4A | 273–341 | 7,768 | 7,768 | 9.1 |
| 3A | 342–400 | 6,664 | 6,665 | 8.2 |
| 2 (C terminal) | 401–416 | 1,875 | 1,876 | 6.8 |
| Subunit (reduced and alkylated) | 48,265 | 48,268 | ||
| Subunit (unmodified)e | 47,173 | 47,030 | 6.4 |
Cloning and sequencing of kamA, the gene for KAM from C. subterminale SB4.
The N-terminal amino acid sequences of KAM and peptide 3A were used to design degenerate oligonucleotides to serve as primers for PCR, which subsequently generated an oligonucleotide of 1,029 bases. Cloning and nucleotide sequence analysis of this fragment yielded approximately 82% of the base sequence of kamA.
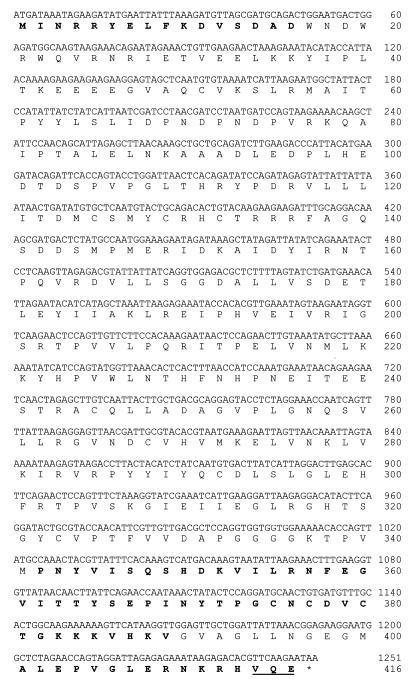
The remaining unknown sequence of kamA was obtained by preparing subgenomic libraries of C. subterminale SB4. A nondegenerate, digoxigenin-containing probe of 500 bp was prepared by PCR. The probe was used to identify _kamA_-containing DNA fragments from restriction-digested clostridial chromosomal DNA. The enzymes were chosen so as to cut within the multicloning region of pUC19 but not within the region of the known incomplete kamA sequence. Subsequent probing showed kamA to reside within chromosomal fragments of 4.3, 4.5, 5.9, and 6.1 kbp for _Xba_I, _Eco_RI, _Acc_I, and _Nde_I digestion, respectively. The start codon, ATG, was found in one _Xba_I colony (X158). The start (ATG) and stop (TAA) codons were found in one _Eco_RI colony (E138). Double-stranded plasmid DNA from these selected colonies was sequenced to obtain the final base sequence of kamA (Fig. 2). Amino acid sequences obtained from N-terminal and C-terminal amino acid analyses of the protein were in perfect agreement with the translated DNA sequence.
FIG. 2.
Nucleotide sequence of kamA and amino acid sequence of C. subterminale SB4 KAM. Sequences in bold are amino acid sequences from N-terminal sequence analyses of the protein and CNBr-treated fragments. The bold and underlined sequence at the C terminus was determined by C-terminal amino acid sequencing.
Correlation of the masses of KAM and CNBr-treated peptides with the gene sequence.
The molecular weight of the translated sequence of amino acids (47,030) agreed with the molecular weight of C. subterminale SB4 KAM obtained by electrospray mass spectrometry (47,173) (Table 1). Additional information obtained from the CNBr-treated peptides is also given in Table 1. HPLC electrospray mass spectrometry of the purified peptides provided information identifying the C-terminal peptide. The final amino acid sequence, based on the translation of the cloned gene described in a later section, included 10 methionine residues instead of the 8 found by amino acid analysis (38). One of the 10 was the N-terminal methionine, which would not yield CNBr-cleaved peptide. In addition, Met 124 and Met 127, as well as Met 145 and Met 147, were so closely spaced that they would yield a tripeptide and dipeptide, respectively, which were not obtained in the purification of peptides. Thus, 8 of the 10 peptides expected based on the translated base sequence were purified and characterized by mass spectrometry (Table 1). The CNBr-treated peptides are arranged in Table 1 in the order in which they appear in the protein. The two very small peptides (molecular mass, <400 Da) were presumably lost in the solvent front during HPLC. Of the eight identified peptides, all had molecular weights equal to the calculated values from the translated base sequence and could be placed in the overall amino acid sequence.
Expression of the C. subterminale SB4 kamA gene in E. coli.
For protein expression, an E. coli expression vector derived from pET23a(+) was prepared. This expression system, when cloned into a cell line which produces an IPTG-inducible T7 RNA polymerase, has been reported to yield very high levels of many heterologous gene products (40). The pAF-80/kamA construct was transformed into competent E. coli BL21(DE3) cells. However, poor expression and poor cell growth were observed with this construct and strain.
Evaluation of the codon usage for the C. subterminale SB4 kamA gene showed that the codon used for all 29 arginine residues is AGA, one of the least frequently used codons in E. coli. From the studies of Kane (22), the expression of heterologous genes containing a high frequency of rare codons (particularly AGG and AGA) in E. coli is difficult or impossible due to low cellular concentrations of their respective tRNA.
It has been suggested that poor expression of the rare AGA codon in E. coli can be relieved by cooverexpression of the E. coli argU gene, which supplies this minor tRNA (6). The sequence of the E. coli argU gene was published by Garcia et al. (16). The primary products of this gene are RNAs of 180 and 190 nucleotides, which are processed in vivo to form the mature arginine tRNA of 77 nucleotides. Therefore, a separate expression vector for argU was prepared. pAlter-Ex2 has a p15a origin of replication that allows it to be maintained with colE1 vectors in the same cell. The presence of this vector confers tetracycline resistance to E. coli.
Serial transformation of E. coli BL21(DE3) cells with both constructs (pAF-80/kamA and pAlter-Ex2/argU) was not absolutely required for the expression of the clostridial kamA gene in E. coli. SDS-PAGE showed a protein band at 47 kDa in whole cells and extracts of cells containing the pAF-80/kamA construct without pAlter-Ex2/argU (data not shown). However, KAM activity of E. coli cellular extracts without pAlter-Ex2/argU was approximately 80% less than that of cells with this construct (Table 2).
TABLE 2.
Activities of recombinant KAM activity in extracts and after purification
| Cloned gene(s) | Sp act (μmol of lysine min−1 mg of protein−1) of: | Yield (mg)a | |
|---|---|---|---|
| Cell extract | Purified enzyme | ||
| None | 0b | ||
| kamA | 1.1 ± 0.2 | 16.7 ± 1.3 | 19 |
| kamA and argU | 5.8 ± 0.5 | 34.5 ± 1.6 | 60 |
| None (native enzyme)c | 35.5 ± 3.1 |
It is worth noting that the expression of kamA in the absence of coexpression of argU severely inhibits cell growth; that is, expression of kamA can be toxic to cells. This was observed in a series of cultures of transformed BL21(DE3) cells. The basal doubling time was 0.8 to 0.9 h for control cells and cells expressing kamA and argU, with or without induction by IPTG. The doubling time for matched cultures expressing kamA without induction was 1.9 h, and for cells expressing kamA with induction by IPTG the doubling time was 5.9 h. Expression of kamA clearly slowed cell growth, and this effect was cured by coexpression of argU.
Both the quantity and quality of the expressed protein were affected by the absence of pAlter-Ex2/argU. The recombinant KAM produced by cells containing pAF-80/kamA and pAlter-Ex2/argU had equivalent enzyme activity (Table 2) and metal content (Fe, 10.0 ± 0.8 g/mol; Zn, 7.6 ± 0.7 g/mol) to the naturally produced clostridial enzyme (25). However, the specific activity of the purified enzyme isolated from cells without pAlter-Ex2/argU was approximately half that of the enzyme isolated from cells containing the argU gene. The yield of purified enzyme from 30 g of cells was also decreased by 65% when argU was absent (Table 2). Expressed in terms of activity units of purified enzyme, coexpression of argU increased the yield of KAM sixfold.
DISCUSSION
The finding of a single N-terminal amino acid sequence and a single C-terminal amino acid sequence for KAM purified from C. subterminale SB4 and the production of fully active KAM by expression of kamA in E. coli prove that KAM consists of a single protomer and that its activity does not depend on any cofactor that cannot be produced by E. coli. It seems safe to conclude that the cofactors SAM and PLP and the [4Fe-4S] center are the only cofactors required for the activity of KAM.
The use of AGA in kamA, a rare codon in E. coli, warranted consideration of a special expression system in E. coli. Earlier accounts indicated that heterologous expression of genes in E. coli that carry this codon in high frequency could be difficult (22). Not only is cell growth and plasmid stability affected by the presence of such genes, but the synthesis of proteins with the correct amino acid sequence may be compromised (6, 22). These problems appear to be caused by a low concentration of the tRNA encoded by argU in E. coli. The present studies showed that expression of kamA in E. coli in the absence of coexpression of the argU gene resulted in slow cell growth and poor expression of KAM. Coexpression of argU with kamA was found to be essential for the practical production of KAM in E. coli. Slow growth and poor expression of KAM in E. coli cannot, however, be attributed solely to the presence of 29 AGA codons in kamA. This is because the genes specifying the subunits for clostridial d-lysine 5,6-aminomutase, an adenosylcobalamin-dependent aminomutase, also incorporate the same rare codon for the arginine residues while nevertheless being reasonably well expressed in E. coli without coexpression of argU (C. H. Chang and P. A. Frey, unpublished data). We note that arginine residues appear in clusters in the structure of KAM at amino acid positions 4 and 5, 25 and 27, and 134, 135, and 136. No such arginine clusters appear in the sequences of the subunits of d-lysine 5,6-aminomutases. A working hypothesis is that the translation of rare codons for clusters of arginine residues in an overexpression system stresses the translation machinery by depleting the availability of the rare species of tRNAArg (AGA), which is required in the production of both KAM and housekeeping proteins in E. coli. For example, the AGA codon is used for arginine in galE, which specifies UDP-galactose 4-epimerase in E. coli. Under this hypothesis, the toxic effect of kamA on cell growth would result from diminished production of housekeeping enzymes.
Identification of proteins with high sequence identity to KAM.
Knowledge of the amino acid sequence of KAM has permitted the search for homologous enzymes in the genomic database. A survey of the available genomic database of translated prokaryotic base sequences revealed seven additional gene products with high sequence identity or similarity to KAM (Fig. 3). Sequence alignments by BestFit revealed identities of 72, 64, 54, 48, 39, 33, and 31% between the amino acid sequence of the clostridial enzyme and the following unknown gene products (GenBank accession number) of Porphyromonas gingivalis (incomplete genome, The Institute for Genomic Research hypothetical protein), Bacillus subtilus (AF015775), Deinococcus radiodurans (incomplete genome, The Institute for Genomic Research hypothetical protein), Aquifex aeolicus (AE000690), Treponema pallidum (AE001197), Haemophilus influenzae (P44641), and E. coli (P39280), respectively. Considering the current sequence information of the reported complete genomes, proteins with amino sequences similar to KAM were not found to be encoded by any of the reported archaeal bacteria nor by Chlamydia trachomatis, Helicobacter pylori, Mycobacterium tuberculosis, and Saccharomyces cerevisiae.
FIG. 3.
Amino acid sequence alignment of KAM with amino acid sequences of gene products of unknown function. From a survey of the available genomic database of translated sequences, amino acid sequences were aligned by PileUp, a computer program of the Genetics Computer Group. C, C. subterminale SB4 KAM; P, P. gingivalis; B, B. subtilis; D, D. radiodurans; A, A. aeolicus; T, T. pallidum; H, H. influenzae; E, E. coli; S, consensus sequence. The numbering in this alignment does not correspond to the numbering of amino acid residues in KAM. All references in the text to amino acid residues in KAM correspond to the numbering in Fig. 2.
The homologous proteins have not been identified. Alignment of the sequences revealed striking similarities with KAM throughout the entire sequence, with few gaps (Fig. 3). The discovery of these genes permits the identification of highly conserved residues. Of the 416 amino acids in KAM, 41 are present in identical positions in the aligned sequences of all eight proteins. There are a total of 190 residues in the consensus sequence. The likelihood that one or more of the homologous proteins are KAMs is significant. In fact, the homologous protein in B. subtilis has been expressed in E. coli and shown to be a KAM (F. J. Ruzicka, D. Chen, and P. A. Frey, unpublished observations).
Clostridial KAM is one of several enzymes that use SAM instead of adenosylcobalamin to generate a putative 5′-deoxyadenosyl radical intermediate. In the case of KAM, this free radical abstracts an unactivated, carbon-bound hydrogen atom and potentiates a 1,2-hydrogen migration with concomitant countermigration of an amino group. ARR activase, PFL activase, and biotin synthase (11, 18, 19, 30, 37, 45) also use SAM to initiate free-radical processes but catalyze different reactions. Amino acid sequence alignments demonstrated little or no homology between KAM and Clostridium pasteurianum or E. coli PFL activase or E. coli ARR activase, apart from the cysteine clusters associated with the iron-sulfur centers common to these proteins (see below). Each alignment showed 22% or less sequence identity to KAM. Not all of the protein components of biotin synthase have been identified and purified to homogeneity. Therefore, comparisons with KAM cannot be made.
Remarkably, short sequences in KAM (320SGYCV324) and its homologs (SGYAV) are similar to the glycine radical motif in PFL, which is 733SGYAV737 in E. coli PFL (44). The glycine radical in ARR appears in a somewhat similar motif (42). No glyine radical has been observed as an intermediate in the reaction of KAM. Moreover, there does not appear to be a role for a glycine radical in the mechanism. This sequence in KAM (amino acids 320 to 324) is near residues 330 to 337, which are postulated below to be part of a SAM binding domain. The apparent glycine radical motif may constitute another part of the SAM binding domain.
The crystal structures and/or amino acid sequences of a large number of SAM binding proteins have been reported. These enzymes include methyltransferases (14, 24, 36) and SAM synthetases (15). SAM is bound in the extended form in a cavity produced by a conserved glycine-rich region that forms a flexible loop (15). Similarly, amino acid residues 330 to 337 (DAPGGGGK) in KAM are highly conserved and may represent a similar region of protein folding required to form a SAM binding domain. The unique chemistry of SAM activation in KAM may forego such a cavity. Distortion at the sulfur-carbon bond of the adenosine 5′ carbon may be necessary to allow for its reductive homolytic cleavage. This in turn may preclude the extended form of SAM. The residues involved in hydrogen bonding or van der Waals contacts (i.e., interactions with adenine N6, 2′-OH of the ribose ring, and the carboxylate oxygens of methionine) are not highly conserved among the various enzymes from different species.
Evaluation of the consensus sequence generated between KAM and the unknown proteins of Fig. 3 indicates that only three cysteines are conserved. These appear in a short group (CXXXCXXC) (Cys125, -129, and -132 in KAM) in all eight of the sequences (Fig. 3). These residues may constitute up to three of four ligands of the iron-sulfur cluster. A similar consensus sequence is found in the activating proteins of E. coli and C. pasteurianum PFL and E. coli ARR (9). The amino acids separating each cysteine residue in this cluster are generally not conserved. However, in the consensus sequence, two neighboring arginine residues, Arg130 and Arg134 in KAM (Arg143 and Arg147 in Fig. 3), are also highly conserved. By providing positive charges, these arginine residues may play an important role in modulating the redox potential of the cluster or nearby SAM molecule, enabling the overall chemistry to occur. A fourth cysteine ligand for the iron-sulfur cluster, if present, is not conserved (9). Inasmuch as we have been unable to prepare an enzyme that contains more than one-half of a [4Fe-4S] cluster per subunit (31), it is possible that only two of the cysteine residues are ligated to the cluster, which is shared between two subunits. Another explanation would suggest a cluster ligated to the protein with only three cysteine residues, the fourth ligation site being occupied by a water or hydroxide ion, as in the case of aconitase (23, 43), or by another amino acid side chain.
A second grouping of cysteine residues appears near the C terminus of KAM in the sequence 375CNCDVC380. KAM contains zinc (31), and the latter cysteine motif may be the zinc binding site. Further experiments will be required to determine whether either cysteine motif constitutes a metal binding site.
The only highly conserved lysine, Lys337 (Lys352 in Fig. 3) is located within the glycine-rich, putative SAM binding domain. It seems unlikely that this conserved lysine would be engaged in an internal aldimine with PLP at the catalytic site. Spectroscopic evidence by electron spin echo envelope modulation spectroscopy has directly implicated the formation of an aldimine linkage of PLP to substrate lysine during catalytic turnover (3). If Lys337 proves not to be part of the SAM binding domain, it may form an internal aldimine with PLP, which may be displaced by the substrate during catalysis. Such a mechanism has been reported for PLP-dependent amino acid aminotransferases (41). It has also been reported that a glycine-rich conserved region (GXXGXXGG) has been found in all known aminolevulinate synthases, glycogen phosphorylase b, and tryptophan synthase (17, 21, 29). In the three-dimensional structures of these PLP-dependent enzymes, this glycine-rich loop associates with the phosphate group of PLP through hydrogen bonding and multiple van der Waals contacts.
In conclusion, putative binding sites for iron and zinc are provided by the amino acid sequence of KAM. A hypothetical SAM binding domain is also present; however, the PLP binding site cannot be assigned on the basis of available information. Practical overexpression of kamA in E. coli requires coexpression of argU, presumably because of the clustering of arginine residues in the amino acid sequence of KAM and the use of AGA as the codon for arginine in the clostridial gene.
ACKNOWLEDGMENTS
We are grateful to Christopher H. Chang for numerous valuable discussions and for his assistance in reading and commenting on this paper.
This research was supported by grant no. DK28607 from the National Institute of Diabetes and Digestive and Kidney Diseases, USPHS.
REFERENCES
- 1.Baker J J, Van der Drift C, Stadtman T C. Purification and properties of β-lysine mutase, a pyridoxal phosphate and B12 coenzyme dependent enzyme. Biochemistry. 1973;12:1054–1063. doi: 10.1021/bi00730a006. [DOI] [PubMed] [Google Scholar]
- 2.Baker J J, Stadtman T C. Aminomutases. In: Dolphin D, editor. B12: biochemistry and medicine. Vol. 2. New York, N.Y: Wiley Interscience; 1982. p. 203. [Google Scholar]
- 3.Ballinger M D, Frey P A, Reed G H, LoBrutto R. Pulsed electron paramagnetic resonance studies of the lysine 2,3-aminomutase substrate radical. Biochemistry. 1995;34:10086–10093. doi: 10.1021/bi00031a033. [DOI] [PubMed] [Google Scholar]
- 4.Ballinger M D, Frey P A, Reed G H. The structure of a substrate radical intermediate in the reaction of lysine 2,3-aminomutase. Biochemistry. 1992;31:10782–10789. doi: 10.1021/bi00159a020. [DOI] [PubMed] [Google Scholar]
- 5.Bjellqvist B, Hughes G J, Pasquali C, Paquet N, Ravier F, Sanchez J C, Frutiger S, Hochstrasser D F. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 6.Brinkmann U, Mattes R E, Buckel P. High level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:109–114. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- 7.Chirpich T P, Zappia V, Costilow R N, Barker H A. Lysine 2,3-aminomutase: purification and properties of a pyridoxal phosphate and S-adenosylmethionine activated enzyme. J Biol Chem. 1970;245:1778–1789. [PubMed] [Google Scholar]
- 8.Costilow R N, Rochovansky O M, Barker H A. Isolation and identification of β-lysine as an intermediate in lysine fermentation. J Biol Chem. 1966;241:1573–1580. [PubMed] [Google Scholar]
- 9.Duin E C, Lafferty M E, Crouse B R, Allen R M, Sanyal I, Flint D H, Johnson M K. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry. 1997;36:11811–11820. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 10.Fontana A, Gross E. Fragmentation of polypeptides by chemical methods. In: Darbre A, editor. Practical protein chemistry. New York, N.Y: Wiley; 1986. pp. 83–88. [Google Scholar]
- 11.Frey M, Rothe M, Volker Wagner A F, Knappe J. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate lyase by abstraction of the glycine C-2 pro-S hydrogen atom. J Biol Chem. 1994;269:12432–12437. [PubMed] [Google Scholar]
- 12.Frey P A, Reed G H. Lysine 2,3-aminomutase and the mechanism of the interconversion of lysine and β-lysine. Adv Enzymol Related Areas Mol Biol. 1993;66:1–39. doi: 10.1002/9780470123126.ch1. [DOI] [PubMed] [Google Scholar]
- 13.Frey P A. Lysine 2,3-aminomutase: is adenosylmethionine a poor man's adenosylcobalamin? FASEB J. 1993;7:662–670. doi: 10.1096/fasebj.7.8.8500691. [DOI] [PubMed] [Google Scholar]
- 14.Fu Z, Hu Y, Konishi K, Takata Y, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry. 1996;35:11985–11993. doi: 10.1021/bi961068n. [DOI] [PubMed] [Google Scholar]
- 15.Fu Z, Hu Y, Markham G D, Takusagawa F. Flexible loop in the structure of S-adenosylmethionine synthetase crystallized in the tetragonal modification. J Biomol Struct Dyn. 1996;13:727–739. doi: 10.1080/07391102.1996.10508887. [DOI] [PubMed] [Google Scholar]
- 16.Garcia G M, Mar P K, Mullin D A, Walker J R, Prather N E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986;45:453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- 17.Gong J, Ferreira G C. Aminolevulinate synthetase: functionally important residues at the glycine loop, a putative pyridoxal phosphate cofactor binding site. Biochemistry. 1995;34:1678–1685. doi: 10.1021/bi00005a024. [DOI] [PubMed] [Google Scholar]
- 18.Guianvarc'h D, Florentin D, Tse Sum Bui B, Nunzi F, Marquet A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem Biophys Res Commun. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 19.Harder J, Eliasson R, Pontis E, Ballinger M D, Reichard P. Activation of the anaerobic ribonucleotide reductase from Escherichia coli by S-adenosylmethionine. J Biol Chem. 1992;267:25548–25552. [PubMed] [Google Scholar]
- 20.Heinrikson R L, Meredith S C. Amino acid analysis by reverse-phase high performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. Anal Biochem. 1984;136:65–74. doi: 10.1016/0003-2697(84)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Hyde C C, Ahmed S A, Padlan E A, Miles E W, Davies D R. Three dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988;263:17857–17871. [PubMed] [Google Scholar]
- 22.Kane J F. Effects of rare codon clusters on high level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy M C, Werst M, Telser J, Emptage M H, Beinert H, Hoffman B M. Mode of substrate carboxyl binding to the [4Fe-4S] cluster of reduced aconitase as studied by 17O and 13C electron nuclear double resonance spectroscopy. Proc Natl Acad Sci USA. 1987;84:8854–8858. doi: 10.1073/pnas.84.24.8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaBahn J, Granzin J, Schluckebier G, Robinson D P, Jack W E, Schildkraut I, Saeger W. Three dimensional structure of the adenine specific DNA methyltransferase MTaqI in complex with the cofactor S-adenosylmethionine. Proc Natl Acad Sci USA. 1994;91:10957–10961. doi: 10.1073/pnas.91.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieder K W, Booker S, Ruzicka F J, Beinert H, Reed G H, Frey P A. S-adenosylmethionine dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance. Biochemistry. 1998;37:2578–2585. doi: 10.1021/bi972417w. [DOI] [PubMed] [Google Scholar]
- 26.Morley C G D, Stadtman T C. Studies on the fermentation of d-alpha lysine. Biochemistry. 1970;9:4890–4900. doi: 10.1021/bi00827a010. [DOI] [PubMed] [Google Scholar]
- 27.Moss M, Frey P A. The role of S-adenosylmethionine in the lysine 2,3-aminomutase reaction. J Biol Chem. 1987;262:14859–14862. [PubMed] [Google Scholar]
- 28.Moss M L, Frey P A. Activation of lysine 2,3-aminomutase by S-adenosylmethionine. J Biol Chem. 1990;265:18112–18115. [PubMed] [Google Scholar]
- 29.Oikonomakos N G, Johnson L N, Acharya K R, Stuart D I, Barford D, Hajdu J, Varvill K M, Melpidou A E, Papageorgiou T, Graves D J, Palm D. Pyridoxal phosphate site in glycogen phosphorylase b: structure in the native enzyme and in three derivatives with modified cofactors. Biochemistry. 1987;26:8381–8389. doi: 10.1021/bi00399a053. [DOI] [PubMed] [Google Scholar]
- 30.Ollagnier S, Mulliez E, Schmidt P P, Eliasson R, Gaillard J, Deronzier C, Bergman T, Graslund A, Reichard P, Fontecave M. Activation of the anaerobic ribonucleotide reductase from Escherichia coli. J Biol Chem. 1997;272:24216–24223. doi: 10.1074/jbc.272.39.24216. [DOI] [PubMed] [Google Scholar]
- 31.Petrovich R M, Ruzicka F J, Reed G H, Frey P A. Characterization of iron-sulfur clusters in lysine 2,3-aminomutase by electron paramagnetic resonance spectroscopy. Biochemistry. 1992;31:10774–10881. doi: 10.1021/bi00159a019. [DOI] [PubMed] [Google Scholar]
- 32.Petrovich R M, Ruzicka F J, Reed G H, Frey P A. Metal cofactors of lysine 2,3-aminomutase. J Biol Chem. 1991;266:7656–7660. [PubMed] [Google Scholar]
- 33.Poston J M. Leucine 2,3-aminomutase, an enzyme of leucine catabolism. J Biol Chem. 1976;251:1859–1863. [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanger F, Niklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schluckebier G, O'Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 37.Shaw N M, Birch O M, Tinschert A, Venetz V, Dietrich R, Savoy L-A. Biotin synthase from Escherichia coli: isolation of an enzyme generated intermediate and stoichiometry of S-adenosylmethionine use. Biochem J. 1998;330:1079–1085. doi: 10.1042/bj3301079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song K B, Frey P A. Molecular properties of lysine 2,3-aminomutase. J Biol Chem. 1991;266:7651–7655. [PubMed] [Google Scholar]
- 39.Stadtman T C. Lysine metabolism by Clostridia. Adv Enzymol Related Areas Mol Biol. 1973;38:413–448. [Google Scholar]
- 40.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 41.Sugio S, Petsko G A, Manning J M, Soda K, Ringe D. Crystal structure of a d-amino acid aminotransferase: how the protein controls stereoselectivity. Biochemistry. 1995;34:9661–9669. doi: 10.1021/bi00030a002. [DOI] [PubMed] [Google Scholar]
- 42.Sun X, Harder J, Krook M, Jornvall H, Sjoberg B-M, Reichard P. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli. Proc Natl Acad Sci USA. 1993;90:577–581. doi: 10.1073/pnas.90.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telser J, Emptage M H, Merkle H, Kennedy M C, Beinert H, Hoffman B M. O-17 electron nuclear double resonance characterization of substrate binding to the [4Fe-4S] cluster of reduced aconitase. J Biol Chem. 1986;261:4840–4846. [PubMed] [Google Scholar]
- 44.Wagner A F, Frey M, Neugebauer F A, Schafer W, Knappe J. The free radical in pyruvate formate lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong K K, Murray B W, Lewisch S A, Baxter M K, Ridky T W, Ulissi-DeMario L, Kozarich J W. Molecular properties of pyruvate formate lyase activating enzyme. Biochemistry. 1993;32:14102–14110. doi: 10.1021/bi00214a005. [DOI] [PubMed] [Google Scholar]