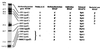Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro - PubMed (original) (raw)
Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro
T J Darcy et al. J Bacteriol. 1999 Jul.
Abstract
RNA polymerase (RNAP) purified from Methanobacterium thermoautotrophicum DeltaH has been shown to initiate transcription accurately in vitro from the hmtB archaeal histone promoter with either native or recombinant forms of the M. thermoautotrophicum TATA-binding protein and transcription factor TFB. Efforts to obtain transcription initiation from hydrogen-regulated methane gene promoters were, however, unsuccessful. Two previously unrecognized archaeal RNAP subunits have been identified, and complex formation by the M. thermoautotrophicum RNAP and TFB has been demonstrated.
Figures
FIG. 1
Identities of M. thermoautotrophicum ΔH RNAP subunits silver stained after separation by tricine-SDS-PAGE. Stetter et al. (24) identified eight subunits (designated by the letters O and A through G) in RNAP preparations from M. thermoautotrophicum Winter. Based on estimated sizes and very similar patterns of SDS-PAGE resolution, O and A through F appear to have been homologs of the M. thermoautotrophicum ΔH subunits here designated A′, B′, B", A", D, E′, and F, and subunit G was a mixture that contained the four polypeptides designated subunits H, L, K, and P. With the exception of subunits F and P, the M. thermoautotrophicum ΔH subunits are homologs of subunits identified previously for Sulfolobus acidocaldarius RNAP (14, 15). M. thermoautotrophicum ΔH RNAP preparations did not contain a homolog of the S. acidocaldarius subunit G, and the MTH0265 and MTH0040 gene products, predicted to be RNAP subunits E" (6.7 kDa) and N (6.4 kDa) (23), were not detected, although their presence may have been masked within the cluster of similarly sized, small subunits. Eucaryal and bacterial homologs of the M. thermoautotrophicum ΔH subunits are listed based on motif conservation and sequence alignments (5, 16, 21, 22). Members of the MTH1324 family (subunit F) have limited similarity to eucaryal Rpb4, a nonessential subunit of RNAPII that in yeast participates in transcription under stress conditions and at temperature extremes (4, 20). All members of the MTH0680.5 family (subunit P) contain four cysteinyl residues arranged in a manner consistent with a C-4 zinc finger motif and homology to Rpb12 (see Fig. 2C). .
FIG. 2
Organization and conservation of the rpoF (A) and rpoP (B) regions in the genomes of M. thermoautotrophicum (MT), A. fulgidus (AF), M. jannaschii (MJ), P. horikoshii (PH), and P. aerophilum (PA) (2, 6, 11, 12, 23). Arrows indicate directions of transcription, shading patterns show homology between genes, and broken lines indicate nonadjacent locations. MTH1323 and MTH0681 and their homologs are predicted to encode rpL21 and rpL37a, respectively. MTH1325 and MTH0680 gene products and their homologs are unknown. Amino acid sequences (C) of MTH0680.5 (MT) and MJ0593.5 (MJ) aligned with their archaeal homologs and the sequences of Rpb12 from Saccharomyces cerevisiae (SC), Schizosaccharomyces pombe (SP), and Homo sapiens (HS) (21, 22). Identical and similar (indicated by asterisks) amino acid residues are identified in the conserved sequence. Four cysteinyl residues predicted to form a C-4 type of zinc finger are boxed. The numbers of amino acid residues present, but not shown, at the N termini of the eucaryal proteins are indicated by the numbers in parentheses. The PH homolog may have longer N-terminal sequences initiated 28 codons upstream at an in-frame ATG (11).
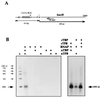
FIG. 3
(A) Template DNA, obtained by _Dde_I digestion of plasmid pRT74 (25), and origin of the 193-nt runoff transcript. (B) Autoradiogram of in vitro-synthesized transcripts. A plus sign indicates that a reaction mixture contained the protein listed. Lane S, size standards; nt, nucleotides.
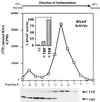
FIG. 4
Sucrose gradient cosedimentation of M. thermoautotrophicum ΔH RNAP and TFB and immunoprecipitation of RNAP by anti-TFB antibodies. Fractions containing TFB and TBP were identified by Western blotting. The inset shows the results of immunoprecipitation experiments with protein A-Sepharose preparations lacking antibodies (−) or coupled to anti-TBP (α-TBP) or anti-TFB (α-TFB). The average values for four separate experiments are shown, with the RNAP activity eluted from each matrix calculated as a percentage of the RNAP activity eluted from the matrix carrying anti-TFB antibodies.
Similar articles
- TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEalpha.
Hanzelka BL, Darcy TJ, Reeve JN. Hanzelka BL, et al. J Bacteriol. 2001 Mar;183(5):1813-8. doi: 10.1128/JB.183.5.1813-1818.2001. J Bacteriol. 2001. PMID: 11160119 Free PMC article. - Identification of a conserved archaeal RNA polymerase subunit contacted by the basal transcription factor TFB.
Magill CP, Jackson SP, Bell SD. Magill CP, et al. J Biol Chem. 2001 Dec 14;276(50):46693-6. doi: 10.1074/jbc.C100567200. Epub 2001 Oct 17. J Biol Chem. 2001. PMID: 11606563 - Determinants of transcription initiation by archaeal RNA polymerase.
Bartlett MS. Bartlett MS. Curr Opin Microbiol. 2005 Dec;8(6):677-84. doi: 10.1016/j.mib.2005.10.016. Epub 2005 Oct 24. Curr Opin Microbiol. 2005. PMID: 16249119 Review. - Archaeal chromatin and transcription.
Reeve JN. Reeve JN. Mol Microbiol. 2003 May;48(3):587-98. doi: 10.1046/j.1365-2958.2003.03439.x. Mol Microbiol. 2003. PMID: 12694606 Review.
Cited by
- Crystal structure of RPB5, a universal eukaryotic RNA polymerase subunit and transcription factor interaction target.
Todone F, Weinzierl RO, Brick P, Onesti S. Todone F, et al. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6306-10. doi: 10.1073/pnas.97.12.6306. Proc Natl Acad Sci U S A. 2000. PMID: 10841537 Free PMC article. - Regulation of tryptophan operon expression in the archaeon Methanothermobacter thermautotrophicus.
Xie Y, Reeve JN. Xie Y, et al. J Bacteriol. 2005 Sep;187(18):6419-29. doi: 10.1128/JB.187.18.6419-6429.2005. J Bacteriol. 2005. PMID: 16159776 Free PMC article. - Redox-sensitive DNA binding by homodimeric Methanosarcina acetivorans MsvR is modulated by cysteine residues.
Isom CE, Turner JL, Lessner DJ, Karr EA. Isom CE, et al. BMC Microbiol. 2013 Jul 16;13:163. doi: 10.1186/1471-2180-13-163. BMC Microbiol. 2013. PMID: 23865844 Free PMC article. - The [4Fe-4S] clusters of Rpo3 are key determinants in the post Rpo3/Rpo11 heterodimer formation of RNA polymerase in Methanosarcina acetivorans.
Jennings ME, Lessner FH, Karr EA, Lessner DJ. Jennings ME, et al. Microbiologyopen. 2017 Feb;6(1):e00399. doi: 10.1002/mbo3.399. Epub 2016 Aug 25. Microbiologyopen. 2017. PMID: 27557794 Free PMC article.
References
- Brown J W, Reeve J N. Transcription initiation and a RNA polymerase binding site upstream of the purE gene of the archaebacterium Methanobacterium thermoautotrophicum strain ΔH. FEMS Microbiol Lett. 1989;60:131–136. - PubMed
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
- Courchesne P L, Luethy R, Patterson S D. Comparison of in-gel and on-membrane digestion methods at low to sub-pmol level for subsequent peptide and fragment-ion mass analysis using matrix-assisted laser-desorption/ionization mass spectrometry. Electrophoresis. 1997;18:369–381. - PubMed
- Edwards A M, Kane C M, Young R A, Kornberg R D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources