LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing - PubMed (original) (raw)
Comparative Study
LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing
Y Kabeya et al. EMBO J. 2000.
Erratum in
- EMBO J. 2003 Sep 1;22(17):4577
Abstract
Little is known about the protein constituents of autophagosome membranes in mammalian cells. Here we demonstrate that the rat microtubule-associated protein 1 light chain 3 (LC3), a homologue of Apg8p essential for autophagy in yeast, is associated to the autophagosome membranes after processing. Two forms of LC3, called LC3-I and -II, were produced post-translationally in various cells. LC3-I is cytosolic, whereas LC3-II is membrane bound. The autophagic vacuole fraction prepared from starved rat liver was enriched with LC3-II. Immunoelectron microscopy on LC3 revealed specific labelling of autophagosome membranes in addition to the cytoplasmic labelling. LC3-II was present both inside and outside of autophagosomes. Mutational analyses suggest that LC3-I is formed by the removal of the C-terminal 22 amino acids from newly synthesized LC3, followed by the conversion of a fraction of LC3-I into LC3-II. The amount of LC3-II is correlated with the extent of autophagosome formation. LC3-II is the first mammalian protein identified that specifically associates with autophagosome membranes.
Figures
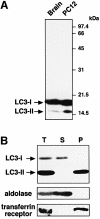
Fig. 1. Two forms of LC3 are produced and found in different locations inside the cells. (A) The lysates of rat brain (left lane) and PC12 cells (right lane) were subjected to immunoblot analysis with the anti-LC3 peptide antibody. (B) A cell homogenate prepared from starved HeLa cells (T) was fractionated into supernatant (S) and pellet (P) by centrifugation at 100 000 g. These fractions were analysed by immunoblot using antibodies against LC3, aldolase (a cytosolic marker) and transferrin receptor (a membrane protein marker). 15 and 12% SDS–polyacrylamide gels were used in (A) and (B), respectively.
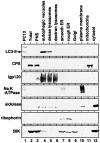
Fig. 2. LC3-II is enriched in autophagic vacuole fractions. Subcellular organelle fractions were obtained from starved rat liver as described in Materials and methods. Each fraction was subjected to immunoblot analysis using antibody against LC3 (top panel) and antibodies against each organelle marker (lower six panels). PC12, total lysate of PC12 cells as a control for the LC3-I and LC3-II positions; Total, total lysate of starved rat liver; PNS, its post-nuclear supernatant. The markers are CPS (carbamoyl phosphate synthetase 1) for the mitochondria; lgp 120 for the endosome/lysosome; Na/K-ATPase for the plasma membrane; aldolase for the cytosol; ribophorin for the ER; and 58 K protein for the Golgi complex.
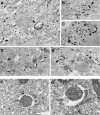
Fig. 3. LC3 is found associated with autophagosome membranes in addition to the cytoplasm. (A–E) HeLa cells transfected with GFP–LC3 were incubated at 37°C for 8 h in the presence of 10 mg/ml HRP and were cultured in Hanks’ solution at 37°C for 60 min. The cells were fixed and stained with DAB to detect the endocytosed HRP. Then, the localization of GFP–LC3 in the cells was examined by silver-enhanced immunogold electron microscopy using antibody against GFP. Arrowheads and arrows in (A–E), respectively, indicate autophagosomes and autolysosomes containing HRP. Bar, 1 µm. (F and G) ES cells were cultured in Hanks’ solution at 37°C for 1 h and fixed. The localization of endogenous LC3 was examined by silver-enhanced immunogold electron microscopy using antibody against LC3. The open and closed arrowheads indicate LC3 associated with inner and outer membranes of the autophagosome, respectively. Bar, 1 µm.
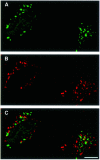
Fig. 4. GFP–LC3 does not co-localize with a lysosomal protein lamp1 in bafilomycin A1-treated cells under starvation conditions. HeLa cells transiently transfected with GFP–LC3 were cultured at 37°C for 90 min in Hanks’ solution containing 0.1 µM bafilomycin A1 and analysed by immunofluorescence confocal microscopy using antibody against lamp1 and rhodamine-conjugated second antibody. (A) GFP–LC3 labelling, (B) lamp1 staining and (C) a merged image of the same field are shown. Bar, 10 µm.
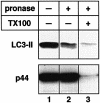
Fig. 5. LC3-II is distributed both outside and inside isolated autophagic vacuoles. Autophagic vacuole fractions were obtained from starved rat liver and were incubated at 0°C for 40 min in the absence (lane 1) or presence (lanes 2 and 3) of 0.8 mg/ml pronase E. In lane 3, 0.2% Triton X-100 was also added. The samples were subjected to immunoblotting with antibodies against LC3 or against BHMT to detect LC3-II and the p44 subunit of BHMT, an autophagic cargo marker.
Fig. 6. Post-translational processing of LC3. (A) Amino acid sequence alignment of the C-terminal segments of LC3 and its homologues. Residues identical to LC3 are shaded in black. Rn, Rattus norvegicus; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Lb, Laccaria bicolor. The sequence of HsLC3 was deduced from an EST clone (au96a10.y1). (B) Constructions of proteins used in (C). Myc epitope tags at the N-terminus, HA epitope tags at the C-terminus and a hypothetical cleavage site Gly120 residue are indicated. To construct LC3G120A, a single point mutation was introduced into LC3, which leads to an amino acid substitution from glycine to alanine at position 120. To construct LC3ΔC22, the 22 C-terminal residues were deleted by PCR. LC3ΔC22,G120A was also produced by site-directed mutagenesis of LC3ΔC22. The ΔC22 mutants were tagged only with the Myc epitopes at the N-teminus. (C) HeLa cells were transiently transfected with Myc-LC3-HA (lanes 1–3 and 13–15), Myc-LC3G120A-HA (lanes 4–6 and 16–18), Myc-LC3ΔC22 (lanes 7–9) or Myc-LC3ΔC22, G120A (lanes 10–12). The cells were labelled with [35S]methionine/cysteine for 4 min and chased for 0, 6 and 90 min at 37°C. The cell lysates were immunoprecipitated with anti-Myc epitope antibody (lanes 1–12) or anti-HA epitope antibody (lanes 13–18) and the immunoprecipitates were analysed by SDS–PAGE and a bioimage analyser.
Fig. 7. The change in the amount of LC3-II corresponds to autophagosome formation. (A) HeLa cells were cultured at 37°C for 90 min in 10% FCS/DMEM (lane 1) or Hanks’ solution (lanes 2–6) containing the following reagents: 1% dimethylsulfoxide (control, lanes 1 and 2); 0.1 µM wortmannin (lane 3); 10 mM 3-methyladenine (lane 4); 0.1 µM bafilomycin A1 (lane 5); and 50 µM vinblastine (lane 6). In a separate experiment, HeLa cells were cultured at 37°C for 120 min in 10% FCS/DMEM (lane 7) or in Hanks’ solution (lane 8), and the latter was followed by re-incubation in 10% FCS/DMEM for 120 min (lane 9). After incubation, the cells were lysed and analysed by immunoblotting using antibody against LC3. A representative experiment repeated twice with duplicated dishes is shown. (B) HeLa cells (open squares with solid lines) and ES cells (open circles with dashed lines) were incubated in Hanks’ solution for the indicated time and a portion of the cells was subjected to immunoblotting using antibody against LC3. The amount of LC3-II was quantified by densitometry (left vertical axis). The remaining HeLa cells were fixed with 2.5% glutaraldehyde for conventional electron microscopy. The area of sections of autophagosomes/autolysosomes was measured on the electron micrographs and is indicated as bars (right vertical axis). (C) HeLa cells transiently transfected with Myc-LC3 were cultured in 10% FCS/DMEM (a) or in Hanks’ solution (b) at 37°C for 90 min. The cells were fixed and permeabilized for immunofluorescence confocal microscopy using anti-Myc epitope antibody and rhodamine-conjugated secondary antibody. Bar, 20 µm.
Similar articles
- Molecular cloning and characterization of rat LC3A and LC3B--two novel markers of autophagosome.
Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y, Pei Y, Wan B, Guo J, Yu L. Wu J, et al. Biochem Biophys Res Commun. 2006 Jan 6;339(1):437-42. doi: 10.1016/j.bbrc.2005.10.211. Epub 2005 Nov 14. Biochem Biophys Res Commun. 2006. PMID: 16300744 - LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation.
Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. Kabeya Y, et al. J Cell Sci. 2004 Jun 1;117(Pt 13):2805-12. doi: 10.1242/jcs.01131. J Cell Sci. 2004. PMID: 15169837 - The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3.
Tanida I, Tanida-Miyake E, Ueno T, Kominami E. Tanida I, et al. J Biol Chem. 2001 Jan 19;276(3):1701-6. doi: 10.1074/jbc.C000752200. Epub 2000 Nov 28. J Biol Chem. 2001. PMID: 11096062 - The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy.
Slobodkin MR, Elazar Z. Slobodkin MR, et al. Essays Biochem. 2013;55:51-64. doi: 10.1042/bse0550051. Essays Biochem. 2013. PMID: 24070471 Review. - Safely removing cell debris with LC3-associated phagocytosis.
Fazeli G, Wehman AM. Fazeli G, et al. Biol Cell. 2017 Oct;109(10):355-363. doi: 10.1111/boc.201700028. Epub 2017 Aug 25. Biol Cell. 2017. PMID: 28755428 Review.
Cited by
- Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes.
Griffin LM, Cicchini L, Pyeon D. Griffin LM, et al. Virology. 2013 Mar 1;437(1):12-9. doi: 10.1016/j.virol.2012.12.004. Epub 2013 Jan 4. Virology. 2013. PMID: 23290079 Free PMC article. - The interplay between autophagy and ROS in tumorigenesis.
Kongara S, Karantza V. Kongara S, et al. Front Oncol. 2012 Nov 21;2:171. doi: 10.3389/fonc.2012.00171. eCollection 2012. Front Oncol. 2012. PMID: 23181220 Free PMC article. - 4-Hydroxytamoxifen induces autophagic death through K-Ras degradation.
Kohli L, Kaza N, Coric T, Byer SJ, Brossier NM, Klocke BJ, Bjornsti MA, Carroll SL, Roth KA. Kohli L, et al. Cancer Res. 2013 Jul 15;73(14):4395-405. doi: 10.1158/0008-5472.CAN-12-3765. Epub 2013 May 30. Cancer Res. 2013. PMID: 23722551 Free PMC article. - Amikacin-induced Fin Reduction is Mediated by Autophagy.
Tsai IT, Chen YH, Chen YH, Wang YH. Tsai IT, et al. J Toxicol Pathol. 2013 Mar;26(1):79-82. doi: 10.1293/tox.26.79. Epub 2013 Apr 22. J Toxicol Pathol. 2013. PMID: 23723573 Free PMC article. - Selective autophagy of AKAP11 activates cAMP/PKA to fuel mitochondrial metabolism and tumor cell growth.
Deng Z, Li X, Blanca Ramirez M, Purtell K, Choi I, Lu JH, Yu Q, Yue Z. Deng Z, et al. Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2020215118. doi: 10.1073/pnas.2020215118. Proc Natl Acad Sci U S A. 2021. PMID: 33785595 Free PMC article.
References
- Blommaart E.F., Luiken,J.J. and Meijer,A.J. (1997) Autophagic proteolysis: control and specificity. Histochem. J., 29, 365–385. - PubMed
- Dunn W.A. (1994) Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol., 4, 139–143. - PubMed
- Hino Y., Asano,A., Sato,R. and Shimizu,S. (1978) Biochemical studies of rat liver Golgi apparatus. I. Isolation and preliminary characterization. J. Biochem. (Tokyo), 83, 909–923. - PubMed
- Hortsch M. and Meyer,D.I. (1985) Immunochemical analysis of rough and smooth microsomes from rat liver. Segregation of docking protein in rough membranes. Eur. J. Biochem., 150, 559–564. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials