Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis - PubMed (original) (raw)
Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis
N L Teterina et al. J Virol. 2001 Apr.
Abstract
HeLa cells were transfected with several plasmids that encoded all poliovirus (PV) nonstructural proteins. Viral RNAs were transcribed by T7 RNA polymerase expressed from recombinant vaccinia virus. All plasmids produced similar amounts of viral proteins that were processed identically; however, RNAs were designed either to serve as templates for replication or to contain mutations predicted to prevent RNA replication. The mutations included substitution of the entire PV 5' noncoding region (NCR) with the encephalomyocarditis virus (EMCV) internal ribosomal entry site, thereby deleting the 5'-terminal cloverleaf-like structure, or insertion of three nucleotides in the 3Dpol coding sequence. Production of viral proteins was sufficient to induce the characteristic reorganization of intracellular membranes into heterogeneous-sized vesicles, independent of RNA replication. The vesicles were stably associated with viral RNA only when RNA replication could occur. Nonreplicating RNAs localized to distinct, nonoverlapping regions in the cell, excluded from the viral protein-membrane complexes. The absence of accumulation of positive-strand RNA from both mutated RNAs in transfected cells was documented. In addition, no minus-strand RNA was produced from the EMCV chimeric template RNA in vitro. These data show that the 5'-terminal sequences of PV RNA are essential for initiation of minus-strand RNA synthesis at its 3' end.
Figures
FIG. 1
(A) Schematic representation of full-length and subgenomic PV RNAs used in this study. PVΔP1 and PVΔP1-3D∗ RNAs contain two nonviral guanylate residues at the 5′ ends of the RNAs transcribed by T7 RNA polymerase. Black bars denote PV sequences; the gray box denotes the EMCV IRES sequence in E5PVΔP1 RNA; the open arrowhead indicates insertion of the codon for Ile in PVΔP1-3D∗; An represents the 30-mer poly(A) tail. (B) Immunoblot analysis of HeLa cells expressing PV proteins. HeLa cells were infected with vTF7-3 and simultaneously transfected with pPVΔP1 (lane 2), pPVΔP1-3D∗ (lane 3), or pE5PVΔP1 (lane 4). Cells were harvested 14 hs after transfection and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting with rabbit anti-PV2C serum (top) or rabbit anti-PV3D serum (bottom). Cells infected with vTF7-3 and mock transfected are shown in lane 1. In all lanes, extracts from approximately 5 × 104 cells were loaded. Positions of 2C, 2BC, 3D, and 3CD (right) and protein markers (left, in kilodaltons) are indicated. (C) In vitro translation of subgenomic RNAs. HeLa cell-free translation reactions were programmed with PV RNA (lane 3) or the indicated subgenomic RNAs transcribed in vitro (lanes 4 to 6). The identities of PV proteins are indicated. Lane 1, marker (M) PV proteins produced in infected cells; lane 2, no RNA.
FIG. 2
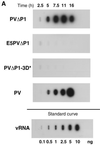
Accumulation of PV-specific RNAs in transfected cells. HeLa cell monolayers were transfected with RNA transcripts in the presence of DEAE-dextran. Cells were harvested at the indicated times after transfection. (A) Accumulation of plus-strand RNA. Total cytoplasmic RNA isolated from approximately 104 cells was bound to a nylon membrane and hybridized to a 32P-labeled riboprobe complementary to nt 5823 to 6056 of the vRNA. The standard curve shows increasing amounts of purified PV RNA (0.1 to 10 ng) hybridized in parallel. (B) Accumulation of negative-strand RNA. Total cytoplasmic RNA isolated from approximately 105 cells was subjected to two-step RNase protection (40). Positions of unprotected (258-nt) and protected (230-nt) probes are indicated. Top, mock-transfected cells (lanes 1 to 5) and cells transfected with PV full-length RNA transcript (lanes 6 to 10); bottom, cells transfected with PVΔP1 (lanes 1 to 5), E5PVΔP1 (lanes 6 to 10), and PVΔP1-3D∗ (lanes 11 to 15) RNA transcripts.
FIG. 2
Accumulation of PV-specific RNAs in transfected cells. HeLa cell monolayers were transfected with RNA transcripts in the presence of DEAE-dextran. Cells were harvested at the indicated times after transfection. (A) Accumulation of plus-strand RNA. Total cytoplasmic RNA isolated from approximately 104 cells was bound to a nylon membrane and hybridized to a 32P-labeled riboprobe complementary to nt 5823 to 6056 of the vRNA. The standard curve shows increasing amounts of purified PV RNA (0.1 to 10 ng) hybridized in parallel. (B) Accumulation of negative-strand RNA. Total cytoplasmic RNA isolated from approximately 105 cells was subjected to two-step RNase protection (40). Positions of unprotected (258-nt) and protected (230-nt) probes are indicated. Top, mock-transfected cells (lanes 1 to 5) and cells transfected with PV full-length RNA transcript (lanes 6 to 10); bottom, cells transfected with PVΔP1 (lanes 1 to 5), E5PVΔP1 (lanes 6 to 10), and PVΔP1-3D∗ (lanes 11 to 15) RNA transcripts.
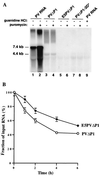
FIG. 3
(A) Synthesis of RNA in vitro. Preinitiation RNA replication complexes were isolated from 40-μl HeLa translation-replication reaction mixtures containing 2 mM guanidine HCl and the indicated RNAs after incubation at 30°C for 6 h. The complexes were resuspended in 40-μl reaction mixtures containing fresh HeLa S10 extract, translation initiation factors, and 25 μCi of [α-32P]CTP. Reaction mixtures 2, 4, 6, and 8 also contained 100 μg of puromycin/ml; reaction mixture 9 contained 2 mM guanidine HCl. The reaction mixtures were incubated at 34°C for 2 h; the labeled RNA products were denatured with glyoxal and characterized by electrophoresis in a 1.1% agarose gel followed by autoradiography (38). The positions of migration of RNA markers run on the same gel and visualized by staining with ethidium bromide are indicated. (B) Stability of RNA templates. 32P-labeled RNAs were synthesized and incubated for various times in HeLa translation-replication reactions, and samples were analyzed on agarose gels as described in Materials and Methods. Intact RNA migrating to the correct position on the gel was quantitated by phosphorimaging. The amounts of radioactivity at the start of the incubation were the same for both samples.
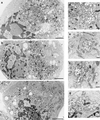
FIG. 4
Electron micrographs of HeLa cells expressing PV nonstructural proteins. Cell cultures were infected with vTF7-3 and transfected with plasmid pPVΔP1 (a and d), pE5PVΔP1 (b and e), or pPVΔP1-3D∗(c and f). (g) PV-infected cell. The morphologies of cells (a to c) and of individual vesicles (d to g) appeared similar in all panels. Bars: 200 (a to c) and 500 (d to g) nm.
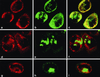
FIG. 5
Localization of viral proteins and RNA in transfected HeLa cells. Cells were infected with vTF7-3 and transfected with pPVΔP1 (a to c), pE5PVΔP1 (d to f), or pPVΔP1-3D∗ (g to i). 2B and 2BC were visualized by IF and CLSM with anti-2B monoclonal antibody and Texas red-labeled secondary antibody (a, d, and g). RNA was localized by FISH with fluorescein isothiocyanate-labeled riboprobe (b, e, and h). Merged images (c, f, and i) show replicating RNA associated with membranes carrying viral nonstructural proteins (c), whereas nonreplicating RNA remains separate (f and i). The horizontal axis of each picture corresponds to 38 μm.
Similar articles
- Inherent instability of poliovirus genomes containing two internal ribosome entry site (IRES) elements supports a role for the IRES in encapsidation.
Johansen LK, Morrow CD. Johansen LK, et al. J Virol. 2000 Sep;74(18):8335-42. doi: 10.1128/jvi.74.18.8335-8342.2000. J Virol. 2000. PMID: 10954532 Free PMC article. - An authentic 3' noncoding region is necessary for efficient poliovirus replication.
Brown DM, Cornell CT, Tran GP, Nguyen JH, Semler BL. Brown DM, et al. J Virol. 2005 Sep;79(18):11962-73. doi: 10.1128/JVI.79.18.11962-11973.2005. J Virol. 2005. PMID: 16140772 Free PMC article. - Genetic analysis of a poliovirus/hepatitis C virus chimera: new structure for domain II of the internal ribosomal entry site of hepatitis C virus.
Zhao WD, Wimmer E. Zhao WD, et al. J Virol. 2001 Apr;75(8):3719-30. doi: 10.1128/JVI.75.8.3719-3730.2001. J Virol. 2001. PMID: 11264361 Free PMC article. - IRES-controlled protein synthesis and genome replication of poliovirus.
Schmid M, Wimmer E. Schmid M, et al. Arch Virol Suppl. 1994;9:279-89. doi: 10.1007/978-3-7091-9326-6_28. Arch Virol Suppl. 1994. PMID: 8032259 Review. - Molecular biology and cell-free synthesis of poliovirus.
Wimmer E, Nomoto A. Wimmer E, et al. Biologicals. 1993 Dec;21(4):349-56. doi: 10.1006/biol.1993.1095. Biologicals. 1993. PMID: 8024750 Review.
Cited by
- Hepatitis A virus infection.
Van Damme P, Pintó RM, Feng Z, Cui F, Gentile A, Shouval D. Van Damme P, et al. Nat Rev Dis Primers. 2023 Sep 28;9(1):51. doi: 10.1038/s41572-023-00461-2. Nat Rev Dis Primers. 2023. PMID: 37770459 Review. - Foot-and-mouth disease virus downregulates vacuolar protein sorting 28 to promote viral replication.
Wang X, Abdullah SW, Wu J, Tang J, Zhang Y, Dong H, Bai M, Wei S, Sun S, Guo H. Wang X, et al. J Virol. 2023 Aug 31;97(8):e0018123. doi: 10.1128/jvi.00181-23. Epub 2023 Aug 11. J Virol. 2023. PMID: 37565750 Free PMC article. - Curcumol inhibits EMCV replication by activating CH25H and inhibiting the formation of ROs.
Zheng J, Sun P, Sun N, Hao Z, Fan K, Yin W, Khan A, Guo J, Zheng X, Li H. Zheng J, et al. BMC Vet Res. 2022 Dec 26;18(1):453. doi: 10.1186/s12917-022-03531-x. BMC Vet Res. 2022. PMID: 36572890 Free PMC article. - How Viruses Use the VCP/p97 ATPase Molecular Machine.
Das P, Dudley JP. Das P, et al. Viruses. 2021 Sep 21;13(9):1881. doi: 10.3390/v13091881. Viruses. 2021. PMID: 34578461 Free PMC article. Review. - Protein Nucleotidylylation in +ssRNA Viruses.
Eruera AR, McSweeney AM, McKenzie-Goldsmith GM, Ward VK. Eruera AR, et al. Viruses. 2021 Aug 5;13(8):1549. doi: 10.3390/v13081549. Viruses. 2021. PMID: 34452414 Free PMC article. Review.
References
- Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources