Porphyromonas gingivalis gingipains and adhesion to epithelial cells - PubMed (original) (raw)
Porphyromonas gingivalis gingipains and adhesion to epithelial cells
T Chen et al. Infect Immun. 2001 May.
Abstract
Porphyromonas gingivalis is one of the principal organisms associated with adult periodontitis. Bacterial surface proteins such as fimbriae and gingipain hemagglutinin domains have been implicated as adhesins that actuate colonization of epithelium lining the gingival sulcus. We investigated the genetics of P. gingivalis adhesion to monolayers of epithelial cells using wild-type and gingipain mutant strains. These experiments suggested that arginine-specific gingipain (Rgp) catalytic activity modulated adhesion. From the data obtained with rgp mutants, we constructed a working hypothesis predicting that attachment and detachment of P. gingivalis to epithelial cells were mediated by gingipain adhesin and Rgp catalytic domains, respectively. A membrane-based epithelial cell binding assay, used to locate adhesins in extracellular fractions of wild-type and mutant strains, recognized gingipain peptides as adhesins rather than fimbriae. We developed a capture assay that demonstrated the binding of gingipain adhesin peptides to oral epithelial cells. The adherence of fimbrillin to epithelial cells was detected after heat denaturation of cell fractions. The prediction that Rgp catalytic activities mediated detachment was substantiated when the high level of attachment of an rgp mutant was reduced in the presence of wild-type cell fractions that contained gingipain catalytic activities.
Figures
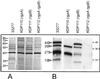
FIG. 1
Structures and homologies of gingipains RgpA, RgpB, Kgp, and HagA. Catalytic domains are depicted as open boxes, and adhesin domains are shaded to indicate specific peptide regions and their homologies. The three repeat regions within HagA from ATCC 33277 were established by PCR (15). The rgpA and kgp genes from ATCC 33277 have not been sequenced; therefore, the assigned molecular masses (kilodaltons) of the peptides are deduced from published sequences of those from the closely related strain 381.
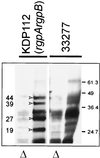
FIG. 2
Gingipain adhesins in VDS fractions from P. gingivalis ATCC 33277 and rgp mutant strains. (A) VDS fractions from parent and mutant strains after SDS-PAGE and silver staining. (B) Western blot of the VDS fractions using a polyclonal antibody to gingipain adhesin domains. The 44-, 39-, 27-, and 19-kDa peptides in the fraction from ATCC 33277 are indicated by arrowheads. The 44-kDa peptide was absent from the rgpA mutant, KDP110. The VDS fraction from the rgpA rgpB mutant, KDP112, did not contain adhesin peptides.
FIG. 3
Western blot of KB epithelial cell extracts after capture of adhesins from VDS fractions of P. gingivalis parent and rgp mutant strains. Before incubation with KB cells, VDS fractions were untreated (−), treated with 500 μM TLCK (T), or heated at 100°C for 5 min (triangles). (A) Blots were probed with antibody to gingipain adhesin domains, and cross-reacting adhesin peptides are indicated by arrowheads. (B) Blots were probed with fimbrillin antibody. Fimbrillin (indicated by the arrowhead) showed more intense reactivity in boiled VDS fractions. WT, wild type.
FIG. 4
N-terminal amino acid sequences of adhesin peptides and sequence comparison of Kgp 44-kDa adhesin peptides. (A) Cleavage sites and N-terminal sequences of the 44-kDa adhesin from RgpA, the 39-kDa adhesin from Kgp, the putative 27-kDa adhesin from Kgp and alternative cleavage site for the 19-kDa peptide, and the 44-kDa adhesins from HagA. (B) Sequence alignment of Kgp 44-kDa adhesins from strains HG66 and 381. N-terminal sequences of the 17- and 27-kDa peptides from HG66 are underlined, and cleavage sites are indicated by arrows. The N terminus of the proposed 27-kDa peptide from 381 is underlined, and alternative downstream processing sites are indicated by arrows.
FIG. 5
Western blot analysis of epithelial cell extracts showing the capture of proteins from cell sonicates of ATCC 33277 and KDP112. Blots were probed with antiadhesin antibody. Triangles denote heat-treated sonicates. The 44-, 39-, 27-, and 19-kDa adhesin peptides (indicated by arrowheads) were present in the cell sonicates of KDP112 and ATCC 33277.
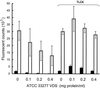
FIG. 6
Adhesion of KDP112 to monolayers is modulated by exogenous gingipain catalytic activities. ATCC 33277 (solid bars) or KDP112 (open bars) was added to KB monolayers in the presence of increasing amounts of the gingipain-containing VDS fraction from ATCC 3277, without or with 500 μM TLCK. Adherent bacteria were detected by their ability to hydrolyze 4-methylumbelliferyl _N_-acetyl-β-
d
-glusosaminide. Bacterial adhesion levels are proportional to 4-methyl umbelliferone fluorescent counts and are expressed as fluorescent counts ± standard deviations.
FIG. 7
Gingipain-mediated adhesion and detachment of P. gingivalis to epithelial cells. We hypothesize that adhesion is mediated by gingipain adhesin peptides localized at the surface of the outer membrane and in membrane vesicles. Rgp catalytic activities, either noncovalently linked with adhesin domains or as soluble proteins, modulate adhesion through digestion of binding substrate. With the parent strain, ATCC 33277, the monolayer adhesion assay measures the net reaction of bacterial attachment to and detachment from receptors or extracellular matrix proteins at the epithelial cell surface. When catalytic activity is reduced by mutation (KDP112), the assay measures the large number of bacteria that attach (via Kgp or HagA adhesin peptides) and accumulate on the monolayers but do not detach or do so very slowly.
Similar articles
- Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells.
Chen T, Duncan MJ. Chen T, et al. Microb Pathog. 2004 Apr;36(4):205-9. doi: 10.1016/j.micpath.2003.12.001. Microb Pathog. 2004. PMID: 15001226 - The role of gingipains in the pathogenesis of periodontal disease.
Imamura T. Imamura T. J Periodontol. 2003 Jan;74(1):111-8. doi: 10.1902/jop.2003.74.1.111. J Periodontol. 2003. PMID: 12593605 Review. - Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis.
Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Nakayama K, et al. J Bacteriol. 1996 May;178(10):2818-24. doi: 10.1128/jb.178.10.2818-2824.1996. J Bacteriol. 1996. PMID: 8631669 Free PMC article. - The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain.
Potempa J, Pike R, Travis J. Potempa J, et al. Infect Immun. 1995 Apr;63(4):1176-82. doi: 10.1128/iai.63.4.1176-1182.1995. Infect Immun. 1995. PMID: 7890369 Free PMC article. - [Structure and functions of trypsin-like cysteine proteinases (gingipains) from Porphyromonas gingivalis, a causative bacterium of periodontitis].
Imamura T. Imamura T. Nihon Saikingaku Zasshi. 2000 Aug;55(3):499-516. Nihon Saikingaku Zasshi. 2000. PMID: 11021086 Review. Japanese. No abstract available.
Cited by
- From Global to Nano: A Geographical Perspective of Aggregatibacter actinomycetemcomitans.
Ryder MI, Fine DH, Barron AE. Ryder MI, et al. Pathogens. 2024 Sep 27;13(10):837. doi: 10.3390/pathogens13100837. Pathogens. 2024. PMID: 39452709 Free PMC article. Review. - Oral microbiota-host interaction: the chief culprit of alveolar bone resorption.
Xu J, Yu L, Ye S, Ye Z, Yang L, Xu X. Xu J, et al. Front Immunol. 2024 Feb 22;15:1254516. doi: 10.3389/fimmu.2024.1254516. eCollection 2024. Front Immunol. 2024. PMID: 38455060 Free PMC article. Review. - A pleiotropic role of sialidase in the pathogenicity of Porphyromonas gingivalis.
Pham C, Guo S, Han X, Coleman L, Sze CW, Wang H, Liu J, Li C. Pham C, et al. Infect Immun. 2024 Mar 12;92(3):e0034423. doi: 10.1128/iai.00344-23. Epub 2024 Feb 20. Infect Immun. 2024. PMID: 38376159 Free PMC article. - A Porphyromonas gingivalis hypothetical protein controlled by the type I-C CRISPR-Cas system is a novel adhesin important in virulence.
Irfan M, Solbiati J, Duran-Pinedo A, Rocha FG, Gibson FC 3rd, Frias-Lopez J. Irfan M, et al. mSystems. 2024 Mar 19;9(3):e0123123. doi: 10.1128/msystems.01231-23. Epub 2024 Feb 7. mSystems. 2024. PMID: 38323815 Free PMC article. - A systematic review of the impact of Porphyromonas gingivalis on foam cell formation: Implications for the role of periodontitis in atherosclerosis.
Afzoon S, Amiri MA, Mohebbi M, Hamedani S, Farshidfar N. Afzoon S, et al. BMC Oral Health. 2023 Jul 13;23(1):481. doi: 10.1186/s12903-023-03183-9. BMC Oral Health. 2023. PMID: 37442956 Free PMC article.
References
- Bhogal P S, Slakeski N, Reynolds E C. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143:2485–2495. - PubMed
- Chen T, Dong H, Yong R, Duncan M J. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb Pathog. 2000;28:235–247. - PubMed
- Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999;34:464–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources