RNA virus error catastrophe: direct molecular test by using ribavirin - PubMed (original) (raw)
RNA virus error catastrophe: direct molecular test by using ribavirin
S Crotty et al. Proc Natl Acad Sci U S A. 2001.
Abstract
RNA viruses evolve rapidly. One source of this ability to rapidly change is the apparently high mutation frequency in RNA virus populations. A high mutation frequency is a central tenet of the quasispecies theory. A corollary of the quasispecies theory postulates that, given their high mutation frequency, animal RNA viruses may be susceptible to error catastrophe, where they undergo a sharp drop in viability after a modest increase in mutation frequency. We recently showed that the important broad-spectrum antiviral drug ribavirin (currently used to treat hepatitis C virus infections, among others) is an RNA virus mutagen, and we proposed that ribavirin's antiviral effect is by forcing RNA viruses into error catastrophe. However, a direct demonstration of error catastrophe has not been made for ribavirin or any RNA virus mutagen. Here we describe a direct demonstration of error catastrophe by using ribavirin as the mutagen and poliovirus as a model RNA virus. We demonstrate that ribavirin's antiviral activity is exerted directly through lethal mutagenesis of the viral genetic material. A 99.3% loss in viral genome infectivity is observed after a single round of virus infection in ribavirin concentrations sufficient to cause a 9.7-fold increase in mutagenesis. Compiling data on both the mutation levels and the specific infectivities of poliovirus genomes produced in the presence of ribavirin, we have constructed a graph of error catastrophe showing that normal poliovirus indeed exists at the edge of viability. These data suggest that RNA virus mutagens may represent a promising new class of antiviral drugs.
Figures
Figure 1
Model of error catastrophe. The majority of viruses in a normal picornavirus population are viable (51). But a small increase in mutation frequency is predicted to push the virus population into error catastrophe (the mutagenized population, Right), where the number of errors per viral genome is sufficiently high to lethally mutate a majority of the virus population. White indicates live virus, gray indicates dead virus. Most animal RNA virus genomes are ≈10 kb long.
Figure 2
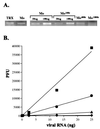
Direct antiviral effect of ribavirin on the viral genetic material. (A) Total poliovirus genomic RNA accumulation in infected cells, with and without ribavirin. On the Left, normal poliovirus genomic RNA (Mo) isolated from infected cells is normalized to a known quantity (100 ng) of in vitro_-generated poliovirus genomes (TRX). On the_Right, quantities of poliovirus genomic RNA isolated from infected cells in 100 μM ribavirin (Mo100r), 400 μM ribavirin (Mo400r), and 1,000 μM ribavirin (Mo1000r), are normalized to amounts of poliovirus genomic RNA from infected untreated cells (Mo). Comparisons are tabulated in Table 2. (B) Large reductions in specific infectivity of ribavirin mutagenized RNA virus genomes. Genomic poliovirus RNA from (■) untreated cells, (●) 100 μM ribavirin-treated cells, (▴) 400 μM ribavirin-treated cells, and (♦) 1,000 μM ribavirin-treated cells was tested for specific infectivity in a series of infectious center assays. Data are shown with a linear curve fit for each series. Assays were performed multiple times; data from a representative experiment is shown.
Figure 3
Distribution of mutations found in the VP1 capsid gene without ribavirin (Δ), with 400 μM ribavirin mutagenesis (*), and 1,000 μM ribavirin mutagenesis (♦). Note that there are no particular mutational hotspots. Almost all mutations at Cs were C→U and mutations at Gs were G→A. Also note that in the presence of 400 μM ribavirin an unusual 3 nt deletion (indicated by —) was detected at a CCA sequence.
Figure 4
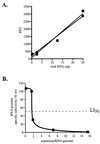
(A) Specific infectivity of T7 transcribed poliovirus genomes compared with natural poliovirus genomes. This illustrates that between 0.21 and 2.1 mutations per 10,000 nt (Table 1), there is no significant detrimental effect on the viability (specific infectivity) of poliovirus genomes. _In vitro_-transcribed RNA (■); natural poliovirus RNA (●). Average data set is shown. (B) Relationship of mutation frequency to genomic RNA infectivity. Specific infectivity of normal poliovirus RNA was set to 100%. The graph shows that poliovirus populations exist near the edge of error catastrophe, as there is a rapid decline in RNA genome infectivity at levels of mutagenesis only slightly higher than normal. The LI50 (50% loss of specific infectivity) is defined as the mutation frequency at which 50% of the viral genomes are lethally mutated, indicated by the dashed line. Wild-type (wt or Mo) poliovirus genomes contain an average ≈1.5 mutations/genome, based on data from Table 1. Poliovirus genomes from cells treated with 100 μM ribavirin (Table 3) contain an average ≈1.9 mutations/genome. Poliovirus genomes from cells treated with 400 μM ribavirin (Table 3) contain an average ≈6.9 mutations/genome. Poliovirus genomes from cells treated with 1,000 μM ribavirin (Table 3) contain an average ≈15.5 mutations/genome.
Similar articles
- Implications of high RNA virus mutation rates: lethal mutagenesis and the antiviral drug ribavirin.
Crotty S, Andino R. Crotty S, et al. Microbes Infect. 2002 Nov;4(13):1301-7. doi: 10.1016/s1286-4579(02)00008-4. Microbes Infect. 2002. PMID: 12443894 Review. - A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity.
Pfeiffer JK, Kirkegaard K. Pfeiffer JK, et al. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7289-94. doi: 10.1073/pnas.1232294100. Epub 2003 May 16. Proc Natl Acad Sci U S A. 2003. PMID: 12754380 Free PMC article. - Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications.
Vignuzzi M, Stone JK, Andino R. Vignuzzi M, et al. Virus Res. 2005 Feb;107(2):173-81. doi: 10.1016/j.virusres.2004.11.007. Virus Res. 2005. PMID: 15649563 Review. - The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen.
Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. Crotty S, et al. Nat Med. 2000 Dec;6(12):1375-9. doi: 10.1038/82191. Nat Med. 2000. PMID: 11100123 - An antiviral mechanism investigated with ribavirin as an RNA virus mutagen for foot-and-mouth disease virus.
Gu CJ, Zheng CY, Zhang Q, Shi LL, Li Y, Qu SF. Gu CJ, et al. J Biochem Mol Biol. 2006 Jan 31;39(1):9-15. doi: 10.5483/bmbrep.2006.39.1.009. J Biochem Mol Biol. 2006. PMID: 16466632
Cited by
- Homology-Based Identification of a Mutation in the Coronavirus RNA-Dependent RNA Polymerase That Confers Resistance to Multiple Mutagens.
Sexton NR, Smith EC, Blanc H, Vignuzzi M, Peersen OB, Denison MR. Sexton NR, et al. J Virol. 2016 Jul 27;90(16):7415-7428. doi: 10.1128/JVI.00080-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27279608 Free PMC article. - On being the right size.
Holmes EC. Holmes EC. Nat Genet. 2005 Sep;37(9):923-4. doi: 10.1038/ng0905-923. Nat Genet. 2005. PMID: 16132047 Free PMC article. - Manganese-dependent polioviruses caused by mutations within the viral polymerase.
Crotty S, Gohara D, Gilligan DK, Karelsky S, Cameron CE, Andino R. Crotty S, et al. J Virol. 2003 May;77(9):5378-88. doi: 10.1128/jvi.77.9.5378-5388.2003. J Virol. 2003. PMID: 12692240 Free PMC article. - Liquid chromatography assay for routine monitoring of cellular ribavirin levels in blood.
Inoue Y, Homma M, Matsuzaki Y, Shibata M, Matsumura T, Ito T, Mitamura K, Tanaka N, Kohda Y. Inoue Y, et al. Antimicrob Agents Chemother. 2004 Oct;48(10):3813-6. doi: 10.1128/AAC.48.10.3813-3816.2004. Antimicrob Agents Chemother. 2004. PMID: 15388439 Free PMC article. Clinical Trial. - Kill or corrupt: Mechanisms of action and drug-resistance of nucleotide analogues against SARS-CoV-2.
Shannon A, Canard B. Shannon A, et al. Antiviral Res. 2023 Feb;210:105501. doi: 10.1016/j.antiviral.2022.105501. Epub 2022 Dec 22. Antiviral Res. 2023. PMID: 36567022 Free PMC article. Review.
References
- Domingo E, Holland J J, Ahlquist P. RNA Genetics. Boca Raton, FL: CRC; 1988.
- Domingo E, Holland J J. In: The Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 161–184.
- Domingo E. Virology. 2000;270:251–253. - PubMed
- Domingo E, Sabo D, Taniguchi T, Weissmann C. Cell. 1978;13:735–744. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources