Short tandem repeat profiling provides an international reference standard for human cell lines - PubMed (original) (raw)
. 2001 Jul 3;98(14):8012-7.
doi: 10.1073/pnas.121616198. Epub 2001 Jun 19.
J A Thomson, B Daly-Burns, Y A Reid, W G Dirks, P Packer, L H Toji, T Ohno, H Tanabe, C F Arlett, L R Kelland, M Harrison, A Virmani, T H Ward, K L Ayres, P G Debenham
Affiliations
- PMID: 11416159
- PMCID: PMC35459
- DOI: 10.1073/pnas.121616198
Short tandem repeat profiling provides an international reference standard for human cell lines
J R Masters et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Cross-contamination between cell lines is a longstanding and frequent cause of scientific misrepresentation. Estimates from national testing services indicate that up to 36% of cell lines are of a different origin or species to that claimed. To test a standard method of cell line authentication, 253 human cell lines from banks and research institutes worldwide were analyzed by short tandem repeat profiling. The short tandem repeat profile is a simple numerical code that is reproducible between laboratories, is inexpensive, and can provide an international reference standard for every cell line. If DNA profiling of cell lines is accepted and demanded internationally, scientific misrepresentation because of cross-contamination can be largely eliminated.
Figures
Figure 1
Fully analyzed data derived from the DNA of cell line HeLaS3, showing the blue, green, and yellow amplified STR peaks and allele classifications. These data are derived from a single lane containing all the PCR products and the red size standards. The PCR products and size standards are detected by laser excitation of the four fluorescent dye labels by using an Applied Biosystems Prism 377 instrument. Profile quality is assessed by two independent analysts, and the peaks are then assigned allele values corresponding to the number of repeat units by using
genescan
and
genotyper
software. Allele designation is based on fixed size windows (±0.5 bp) derived from multiple analyses of allelic ladder samples containing all the commonly occurring alleles (data not shown). The loci are as follows: blue (Left to Right): HUMAMGX/Y, HUMTH01, D21S11, D18S51; green: D8S1179; yellow: HUMVWFA31/A, HUMFIBRA.
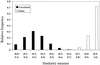
Figure 2
Histogram showing relative frequency of the similarity measure at 10% intervals for the 6,903 unrelated pairs of cell lines (black bars) and 264 pairs of cell lines of the same origin (open bars).
Comment in
- Cell culture forensics.
O'Brien SJ. O'Brien SJ. Proc Natl Acad Sci U S A. 2001 Jul 3;98(14):7656-8. doi: 10.1073/pnas.141237598. Proc Natl Acad Sci U S A. 2001. PMID: 11438719 Free PMC article. No abstract available.
Similar articles
- Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines.
Dirks WG, Faehnrich S, Estella IA, Drexler HG. Dirks WG, et al. ALTEX. 2005;22(2):103-9. ALTEX. 2005. PMID: 15953965 - Essential role for gene profiling analysis in the authentication of human cell lines.
Yoshino K, Iimura E, Saijo K, Iwase S, Fukami K, Ohno T, Obata Y, Nakamura Y. Yoshino K, et al. Hum Cell. 2006 Feb;19(1):43-8. doi: 10.1111/j.1749-0774.2005.00007.x. Hum Cell. 2006. PMID: 16643607 - Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues.
Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, Furtado M, Kline MC, Kohara A, Los GV, MacLeod RA, Masters JR, Nardone M, Nardone RM, Nims RW, Price PJ, Reid YA, Shewale J, Sykes G, Steuer AF, Storts DR, Thomson J, Taraporewala Z, Alston-Roberts C, Kerrigan L. Barallon R, et al. In Vitro Cell Dev Biol Anim. 2010 Oct;46(9):727-32. doi: 10.1007/s11626-010-9333-z. Epub 2010 Jul 8. In Vitro Cell Dev Biol Anim. 2010. PMID: 20614197 Free PMC article. - Authentication of human cell-based products: the role of a new consensus standard.
Kerrigan L, Nims RW. Kerrigan L, et al. Regen Med. 2011 Mar;6(2):255-60. doi: 10.2217/rme.11.5. Regen Med. 2011. PMID: 21391858 Review. - Cell line misidentification: the beginning of the end.
American Type Culture Collection Standards Development Organization Workgroup ASN-0002. American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Nat Rev Cancer. 2010 Jun;10(6):441-8. doi: 10.1038/nrc2852. Epub 2010 May 7. Nat Rev Cancer. 2010. PMID: 20448633 Review.
Cited by
- Synergistic activity of rapamycin and dexamethasone in vitro and in vivo in acute lymphoblastic leukemia via cell-cycle arrest and apoptosis.
Zhang C, Ryu YK, Chen TZ, Hall CP, Webster DR, Kang MH. Zhang C, et al. Leuk Res. 2012 Mar;36(3):342-9. doi: 10.1016/j.leukres.2011.10.022. Epub 2011 Dec 3. Leuk Res. 2012. PMID: 22137317 Free PMC article. - Multiplex short tandem repeat profiling of immortalized hepatic stellate cell line Col-GFP HSC.
Meurer SK, Brenner DA, Weiskirchen R. Meurer SK, et al. PLoS One. 2022 Sep 6;17(9):e0274219. doi: 10.1371/journal.pone.0274219. eCollection 2022. PLoS One. 2022. PMID: 36067186 Free PMC article. - iTIME.219: An Immortalized KSHV Infected Endothelial Cell Line Inducible by a KSHV-Specific Stimulus to Transition From Latency to Lytic Replication and Infectious Virus Release.
Dollery SJ, Maldonado TD, Brenner EA, Berger EA. Dollery SJ, et al. Front Cell Infect Microbiol. 2021 Apr 14;11:654396. doi: 10.3389/fcimb.2021.654396. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33937098 Free PMC article. - Therapeutic potential for phenytoin: targeting Na(v)1.5 sodium channels to reduce migration and invasion in metastatic breast cancer.
Yang M, Kozminski DJ, Wold LA, Modak R, Calhoun JD, Isom LL, Brackenbury WJ. Yang M, et al. Breast Cancer Res Treat. 2012 Jul;134(2):603-15. doi: 10.1007/s10549-012-2102-9. Epub 2012 Jun 8. Breast Cancer Res Treat. 2012. PMID: 22678159 Free PMC article. - Verification and unmasking of widely used human esophageal adenocarcinoma cell lines.
Boonstra JJ, van Marion R, Beer DG, Lin L, Chaves P, Ribeiro C, Pereira AD, Roque L, Darnton SJ, Altorki NK, Schrump DS, Klimstra DS, Tang LH, Eshleman JR, Alvarez H, Shimada Y, van Dekken H, Tilanus HW, Dinjens WN. Boonstra JJ, et al. J Natl Cancer Inst. 2010 Feb 24;102(4):271-4. doi: 10.1093/jnci/djp499. Epub 2010 Jan 14. J Natl Cancer Inst. 2010. PMID: 20075370 Free PMC article.
References
- Stacey G N, Masters J R W, Hay R J, Drexler H G, MacLeod R A F, Freshney R I. Nature (London) 2000;403:356. - PubMed
- Gey G O, Coffman W D, Kubicek M T. Cancer Res. 1952;12:264–265.
- Defendi V, Billingham R E, Silvers W K, Moorhead P. J Natl Cancer Inst. 1960;25:359–385. - PubMed
- Brand K G, Syverton J T. J Natl Cancer Inst. 1962;28:147–157. - PubMed
- Gartler S M. Natl Cancer Inst Monogr. 1967;26:167–195. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials