A conditional tissue-specific transgene expression system using inducible GAL4 - PubMed (original) (raw)
A conditional tissue-specific transgene expression system using inducible GAL4
T Osterwalder et al. Proc Natl Acad Sci U S A. 2001.
Abstract
In Drosophila, the most widely used system for generating spatially restricted transgene expression is based on the yeast GAL4 protein and its target upstream activating sequence (UAS). To permit temporal as well as spatial control over UAS-transgene expression, we have explored the use of a conditional RU486-dependent GAL4 protein (GeneSwitch) in Drosophila. By using cloned promoter fragments of the embryonic lethal abnormal vision gene or the myosin heavy chain gene, we have expressed GeneSwitch specifically in neurons or muscles and show that its transcriptional activity within the target tissues depends on the presence of the activator RU486 (mifepristone). We used available UAS-reporter lines to demonstrate RU486-dependent tissue-specific transgene expression in larvae. Reporter protein expression could be detected 5 h after systemic application of RU486 by either feeding or "larval bathing." Transgene expression levels were dose-dependent on RU486 concentration in larval food, with low background expression in the absence of RU486. By using genetically altered ion channels as reporters, we were able to change the physiological properties of larval bodywall muscles in an RU486-dependent fashion. We demonstrate here the applicability of GeneSwitch for conditional tissue-specific expression in Drosophila, and we provide tools to control pre- and postsynaptic expression of transgenes at the larval neuromuscular junction during postembryonic life.
Figures
Figure 1
The GeneSwitch/UAS expression system in Drosophila. Driver lines expressing the transcriptional activator GeneSwitch in a tissue-specific fashion are crossed to UAS-reporter lines with genomic inserts of a target gene fused to five GAL4-binding sites arrayed in tandem (5× UAS). In the absence of an activator, the GeneSwitch protein is expressed in target tissues but remains transcriptionally silent (black); Gene X is therefore not expressed. However, after systemic application of RU486 (red), the GeneSwitch protein becomes transcriptionally active (blue), mediating expression of gene X (green) in only those tissues expressing GeneSwitch. [Reproduced with permission from ref. (Copyright 1993, The Company of Bioligists Limited).]
Figure 2
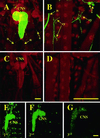
RU486 activates GeneSwitch in the larval nervous system. (A–D) Confocal images of fixed bodywall preparations of Drosophila third instar larvae expressing cytosolic GFP (green channel) from a neuron-specific GeneSwitch driver. Larvae were raised in the presence (A, B) or absence (C, D) of RU486. Counterlabeling is for the nuclear localized even skipped (red channel). (E–G) Confocal images of unfixed, whole-mount first (E), second (F), or third (G) instar larvae expressing UAS-EGFP from ELAV-GeneSwitch. GeneSwitch was activated in embryos by feeding mothers RU486, but first instar larvae were transferred to, and developed on, normal food. (CNS, central nervous system; s, sensory neurons; n, nerves; sy, neuromuscular synapses.) (Bar = 100 μm.)
Figure 3
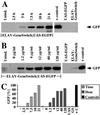
RU486-mediated expression starts rapidly and is dose-dependent. Quantitative Western blot analysis of GFP protein. Genotypes of the larvae are ELAV-GeneSwitch; UAS-EGFP if not otherwise indicated. UAS-EGFP;+ (UAS) and +;ELAV-GeneSwitch (GS) are parental lines; ELAV-GAL4;UAS-EGFP animals have constitutively active GAL4 in all neurons serve as a positive control (+ control, +C). (A) Timecourse of GFP protein (arrow) expression (hours after “larval bathing”) and (B) dose dependence of GFP protein expression on RU486 concentration (μg/ml) in larval food. (C) Qantitative analysis of Western blots in A (light gray, “Time”) and B (dark gray, “Dose”) normalized to positive control (ELAV-GAL4;UAS-EGFP, black).
Figure 4
Both basal transcriptional activity of ELAV-GeneSwitch and toxicity of RU486 are low. The viability of embryos expressing either a UAS-TNTE reporter (black bars, TnTx[e]) or a control UAS-EGFP reporter (gray bars, EGFP) from the ELAV-GeneSwitch driver is shown as a function of RU486 concentrations in the parents' food. The percentage of UAS-TNTE/ELAV-GeneSwitch first instar larvae surviving to adulthood in the absence of RU486 is shown as a hatched bar (TnTx[a]).
Figure 5
RU486 induces transgene expression in larval bodywall muscles. (A) Expression of UAS-EGFP driven by MHC-GeneSwitch in bodywall muscles of first (Left), early third (Center), and late third instar larvae (Right) in the presence (Bottom) or absence (Top) of RU486. The bodywall musculature was imaged through the cuticle in undissected larvae. Muscle fibers 7, 6, 13, 12, and 5 are labeled; first instar larva or muscles are outlined in red in uninduced animals for better visibility. (B) Schematic diagram for two electrode voltage-clamp analyses (Upper) and EKO-channel localization predominantly to NMJs in fixed bodywall preparations from third instar UAS-EKO/MHC-GeneSwitch larvae (Lower). (C) Representative current traces and (D) current–voltage relationship obtained under zero Ca2+ conditions for uninduced (blue traces and curves) or induced (red traces and curves) UAS-EKO/MHC-GeneSwitch animals (left column in C, squares in D) or UAS-EGFP/MHC-GeneSwitch controls (right column in C, circles in D).
Similar articles
- Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers.
Nicholson L, Singh GK, Osterwalder T, Roman GW, Davis RL, Keshishian H. Nicholson L, et al. Genetics. 2008 Jan;178(1):215-34. doi: 10.1534/genetics.107.081968. Genetics. 2008. PMID: 18202369 Free PMC article. - The RU486-dependent activation of the GeneSwitch system in adult muscles leads to severe adverse effects in Drosophila.
Zappia MP, Damschroder D, Westacott A, Wessells RJ, Frolov MV. Zappia MP, et al. G3 (Bethesda). 2024 May 7;14(5):jkae039. doi: 10.1093/g3journal/jkae039. G3 (Bethesda). 2024. PMID: 38409337 Free PMC article. - A hormone receptor-based transactivator bridges different binary systems to precisely control spatial-temporal gene expression in Drosophila.
Kuo SY, Tu CH, Hsu YT, Wang HD, Wen RK, Lin CT, Wu CL, Huang YT, Huang GS, Lan TH, Fu TF. Kuo SY, et al. PLoS One. 2012;7(12):e50855. doi: 10.1371/journal.pone.0050855. Epub 2012 Dec 11. PLoS One. 2012. PMID: 23239992 Free PMC article. - The GAL4 system : a versatile system for the expression of genes.
Elliott DA, Brand AH. Elliott DA, et al. Methods Mol Biol. 2008;420:79-95. doi: 10.1007/978-1-59745-583-1_5. Methods Mol Biol. 2008. PMID: 18641942 Review. - Ectopic gene expression in Drosophila using GAL4 system.
Phelps CB, Brand AH. Phelps CB, et al. Methods. 1998 Apr;14(4):367-79. doi: 10.1006/meth.1998.0592. Methods. 1998. PMID: 9608508 Review.
Cited by
- Lack of miRNA Misregulation at Early Pathological Stages in Drosophila Neurodegenerative Disease Models.
Reinhardt A, Feuillette S, Cassar M, Callens C, Thomassin H, Birman S, Lecourtois M, Antoniewski C, Tricoire H. Reinhardt A, et al. Front Genet. 2012 Oct 30;3:226. doi: 10.3389/fgene.2012.00226. eCollection 2012. Front Genet. 2012. PMID: 23115562 Free PMC article. - Independent glial subtypes delay development and extend healthy lifespan upon reduced insulin-PI3K signalling.
Woodling NS, Rajasingam A, Minkley LJ, Rizzo A, Partridge L. Woodling NS, et al. BMC Biol. 2020 Sep 14;18(1):124. doi: 10.1186/s12915-020-00854-9. BMC Biol. 2020. PMID: 32928209 Free PMC article. - Traumatic injury compromises nucleocytoplasmic transport and leads to TDP-43 pathology.
Anderson EN, Morera AA, Kour S, Cherry JD, Ramesh N, Gleixner A, Schwartz JC, Ebmeier C, Old W, Donnelly CJ, Cheng JP, Kline AE, Kofler J, Stein TD, Pandey UB. Anderson EN, et al. Elife. 2021 May 26;10:e67587. doi: 10.7554/eLife.67587. Elife. 2021. PMID: 34060470 Free PMC article. - Genetic and metabolomic architecture of variation in diet restriction-mediated lifespan extension in Drosophila.
Jin K, Wilson KA, Beck JN, Nelson CS, Brownridge GW 3rd, Harrison BR, Djukovic D, Raftery D, Brem RB, Yu S, Drton M, Shojaie A, Kapahi P, Promislow D. Jin K, et al. PLoS Genet. 2020 Jul 9;16(7):e1008835. doi: 10.1371/journal.pgen.1008835. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32644988 Free PMC article. - Stress granule formation helps to mitigate neurodegeneration.
Glineburg MR, Yildirim E, Gomez N, Li X, Pak J, Altheim C, Waksmacki J, McInerney G, Barmada SJ, Todd PK. Glineburg MR, et al. bioRxiv [Preprint]. 2023 Nov 11:2023.11.07.566060. doi: 10.1101/2023.11.07.566060. bioRxiv. 2023. PMID: 37986813 Free PMC article. Updated. Preprint.
References
- Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. - PubMed
- Luo L, Liao Y J, Jan L Y, Jan Y N. Genes Dev. 1994;8:1787–1802. - PubMed
- Gustafson K, Boulianne G L. Genome. 1996;39:174–182. - PubMed
- Brand A H, Dormand E L. Curr Opin Neurobiol. 1995;5:572–578. - PubMed
- Phelps C B, Brand A H. Methods. 1998;14:367–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous