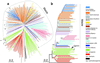A single domestication for maize shown by multilocus microsatellite genotyping - PubMed (original) (raw)
A single domestication for maize shown by multilocus microsatellite genotyping
Yoshihiro Matsuoka et al. Proc Natl Acad Sci U S A. 2002.
Abstract
There exists extraordinary morphological and genetic diversity among the maize landraces that have been developed by pre-Columbian cultivators. To explain this high level of diversity in maize, several authors have proposed that maize landraces were the products of multiple independent domestications from their wild relative (teosinte). We present phylogenetic analyses based on 264 individual plants, each genotyped at 99 microsatellites, that challenge the multiple-origins hypothesis. Instead, our results indicate that all maize arose from a single domestication in southern Mexico about 9,000 years ago. Our analyses also indicate that the oldest surviving maize types are those of the Mexican highlands with maize spreading from this region over the Americas along two major paths. Our phylogenetic work is consistent with a model based on the archaeological record suggesting that maize diversified in the highlands of Mexico before spreading to the lowlands. We also found only modest evidence for postdomestication gene flow from teosinte into maize.
Figures
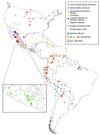
Figure 1
Geographic distribution of maize and teosinte used in this study. Core Andean maize characterized by hand-grenade-shaped ears (22 samples), other South American maize (47), Guatemalan and southern Mexican maize (31), Caribbean maize (6), lowland western and northern Mexican maize (15), highland Mexican maize (20), eastern and central U.S. maize (24), southwestern U.S. maize (22), northern Mexican maize (6), ssp. parviglumis (34), and ssp. mexicana (33). Inset shows the distribution of the 34 populations of ssp. parviglumis in southern Mexico with the populations that are basal to maize in Fig. 2 (represented as asterisks). The blue line is the Balsas River and its major tributaries.
Figure 2
Phylogenies of maize and teosinte rooted with ssp. huehuetenangensis based on 99 microsatellites. Dashed gray line circumscribes the monophyletic maize lineage. Asterisks identify those populations of ssp. parviglumis basal to maize, all of which are from the central Balsas River drainage. (a) Individual plant tree based on 193 maize and 71 teosinte. (b) Tree based on 95 ecogeographically defined groups. The numbers on the branches indicate the number of times a clade appeared among 1,000 bootstrap samples. Only bootstrap values greater than 900 are shown. The arrow indicates the position of Oaxacan highland maize that is basal to all of the other maize.
Figure 3
Graph of the first two axes from a principal component analysis of 193 maize and 71 teosinte individual plants. The first component explains 3.5% and the second 2.6% of the total variation.
Similar articles
- Domestication and lowland adaptation of coastal preceramic maize from Paredones, Peru.
Vallebueno-Estrada M, Hernández-Robles GG, González-Orozco E, Lopez-Valdivia I, Rosales Tham T, Vásquez Sánchez V, Swarts K, Dillehay TD, Vielle-Calzada JP, Montiel R. Vallebueno-Estrada M, et al. Elife. 2023 Apr 18;12:e83149. doi: 10.7554/eLife.83149. Elife. 2023. PMID: 37070964 Free PMC article. - Genetic diversity and population structure of teosinte.
Fukunaga K, Hill J, Vigouroux Y, Matsuoka Y, Sanchez G J, Liu K, Buckler ES, Doebley J. Fukunaga K, et al. Genetics. 2005 Apr;169(4):2241-54. doi: 10.1534/genetics.104.031393. Epub 2005 Jan 31. Genetics. 2005. PMID: 15687282 Free PMC article. - Evidence of selection at the ramosa1 locus during maize domestication.
Sigmon B, Vollbrecht E. Sigmon B, et al. Mol Ecol. 2010 Apr;19(7):1296-311. doi: 10.1111/j.1365-294X.2010.04562.x. Epub 2010 Feb 24. Mol Ecol. 2010. PMID: 20196812 - Genomic screening for artificial selection during domestication and improvement in maize.
Yamasaki M, Wright SI, McMullen MD. Yamasaki M, et al. Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review. - The genetics of maize evolution.
Doebley J. Doebley J. Annu Rev Genet. 2004;38:37-59. doi: 10.1146/annurev.genet.38.072902.092425. Annu Rev Genet. 2004. PMID: 15568971 Review.
Cited by
- Genome size variation in wild and cultivated maize along altitudinal gradients.
Díez CM, Gaut BS, Meca E, Scheinvar E, Montes-Hernandez S, Eguiarte LE, Tenaillon MI. Díez CM, et al. New Phytol. 2013 Jul;199(1):264-276. doi: 10.1111/nph.12247. Epub 2013 Apr 2. New Phytol. 2013. PMID: 23550586 Free PMC article. - Asymmetrical local adaptation of maize landraces along an altitudinal gradient.
Mercer K, Martínez-Vásquez Á, Perales HR. Mercer K, et al. Evol Appl. 2008 Aug;1(3):489-500. doi: 10.1111/j.1752-4571.2008.00038.x. Evol Appl. 2008. PMID: 25567730 Free PMC article. - Comparative transcriptomics uncovers alternative splicing changes and signatures of selection from maize improvement.
Huang J, Gao Y, Jia H, Liu L, Zhang D, Zhang Z. Huang J, et al. BMC Genomics. 2015 May 8;16(1):363. doi: 10.1186/s12864-015-1582-5. BMC Genomics. 2015. PMID: 25952680 Free PMC article. - Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time.
Wang Y, Gu Y, Gao H, Qiu L, Chang R, Chen S, He C. Wang Y, et al. BMC Evol Biol. 2016 Apr 12;16:79. doi: 10.1186/s12862-016-0653-9. BMC Evol Biol. 2016. PMID: 27072125 Free PMC article. - Origins of maize: a further paradox resolved.
Sawers RJ, Sanchez Leon NL. Sawers RJ, et al. Front Genet. 2011 Aug 23;2:53. doi: 10.3389/fgene.2011.00053. eCollection 2011. Front Genet. 2011. PMID: 22303349 Free PMC article. No abstract available.
References
- Second G. Jpn J Genet. 1982;57:25–57.
- Sonnante G, Stockton T, Nodari R O, Becerra Velásquez V L, Gepts P. Theor Appl Genet. 1994;89:629–635. - PubMed
- Yabuno T. Cytologia. 1962;27:296–305.
- Wendel J F. In: Evolution of Crop Plants. Smartt J, Simmonds N W, editors. Essex, U.K.: Longman; 1995. pp. 358–366.
- Decker D S. Econ Bot. 1988;42:4–15.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources