Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae - PubMed (original) (raw)
Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae
Cecilia J Bonangelino et al. Mol Biol Cell. 2002 Jul.
Abstract
The biosynthetic sorting of hydrolases to the yeast vacuole involves transport along two distinct routes referred to as the carboxypeptidase Y and alkaline phosphatase pathways. To identify genes involved in sorting to the vacuole, we conducted a genome-wide screen of 4653 homozygous diploid gene deletion strains of Saccharomyces cerevisiae for missorting of carboxypeptidase Y. We identified 146 mutant strains that secreted strong-to-moderate levels of carboxypeptidase Y. Of these, only 53 of the corresponding genes had been previously implicated in vacuolar protein sorting, whereas the remaining 93 had either been identified in screens for other cellular processes or were only known as hypothetical open reading frames. Among these 93 were genes encoding: 1) the Ras-like GTP-binding proteins Arl1p and Arl3p, 2) actin-related proteins such as Arp5p and Arp6p, 3) the monensin and brefeldin A hypersensitivity proteins Mon1p and Mon2p, and 4) 15 novel proteins designated Vps61p-Vps75p. Most of the novel gene products were involved only in the carboxypeptidase Y pathway, whereas a few, including Mon1p, Mon2p, Vps61p, and Vps67p, appeared to be involved in both the carboxypeptidase Y and alkaline phosphatase pathways. Mutants lacking some of the novel gene products, including Arp5p, Arp6p, Vps64p, and Vps67p, were severely defective in secretion of mature alpha-factor. Others, such as Vps61p, Vps64p, and Vps67p, displayed defects in the actin cytoskeleton at 30 degrees C. The identification and phenotypic characterization of these novel mutants provide new insights into the mechanisms of vacuolar protein sorting, most notably the probable involvement of the actin cytoskeleton in this process.
Figures
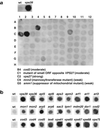
Figure 1
Identification of CPY-secreting strains. The collection of 4653 homozygous diploid deletion strains developed by the Saccharomyces Genome Deletion Project was screened using a CPY colony blot assay (Roberts et al., 1991). The membranes were subjected to immunoblotting with mouse anti-CPY at 1:1000 dilution. Results for either one 96-well plate (a) or examples of several mutants taken from different blots (b) are shown. Wild-type (wt) and vps39 strains are included for comparison.
Figure 2
Analysis of CPY, PrA, and ALP processing. Wild-type (wt) and deletion strains were pulse-labeled with [35S]methionine for 10 min (first lane of each pair) and chased for 10 min (second lane of each pair). Sequential immunoprecipitations were performed from the lysates by using antibodies against CPY (a), PrA (b), or ALP (c) as described in MATERIALS AND METHODS. Results are shown for wild-type (wt) and 24 mutant strains. The positions of precursor (p) and mature (m) forms of the hydrolases examined are indicated. The p1 and p2 precursor forms of CPY were not well resolved for all strains, in some cases due to aberrant Golgi glycosylation that resulted in comigration of both precursor species. A summary of results for these and other strains not shown in this figure is presented in Table 3.
Figure 3
Visualization of vacuole morphology in wild-type and mutant strains by light microscopy. Vacuole morphology was examined by labeling with FM4-64 as described in MATERIALS AND METHODS. The photographs were taken with both fluorescence and a low level of transmitted light. Results are shown for wild-type (wt), vps39 (as a control for aberrant vacuole morphology), and 23 selected vacuolar sorting mutants identified in our screen. Many of the deletions resulted in fragmented vacuole morphology, which was intermediate, and not as severe as the class B vps mutants (vps39). These include arf1, arl1, arl3, mon1, mon2, mdm20, tpm1, dor1, cod2, cod3, vps61, and vps67. Bar, 9 μm.
Figure 4
Visualization of the actin cytoskeleton in wild-type and mutant strains by light microscopy. The actin cytoskeleton was examined by labeling fixed log-phase cells with 1.1 μM Oregon green-phalloidin at 30°C for ∼16–20 h as described in MATERIALS AND METHODS. Results are shown for wild-type (wt), vps39, and 23 selected vacuolar sorting mutants. The most common phenotype observed was the decrease in number or an absence of polarized actin cable in the mother cells, as is seen in arf1, mdm20, tpm1, and vps61. Other phenotypes included reduced polarized actin cables with the appearance of a large actin aggregate in the mother cell, seen in vps36 and vps65 (arrowheads), and slightly disorganized actin filaments, observed in arp5 and arp6. Bar, 9 μm.
Figure 5
Analysis of α-factor secretion and processing. (a) To test for α-factor secretion, wild-type and mutant strains were grown to log phase and serially diluted to the concentrations indicated, and equal amounts were spotted on YEPD plates containing a freshly spread lawn of the α-factor–sensitive strain RC634 (MAT a, sst1-3), as described in MATERIALS AND METHODS. The plates were then incubated at 30°C for 48 h to assess α-factor secretion as indicated by the relative growth inhibition of the sst1-3 mutant strain (halo). (b) To test for processing of α-factor, strains were metabolically labeled with [35S]methionine at 25°C for 7.5 min as described in MATERIALS AND METHODS. Mature α-factor (mαf), α-factor–processing intermediates and pro-α-factor were immunoprecipitated with anti-α-factor antibodies, and resolved by SDS-PAGE (4–20% acrylamide gradient gels). Various forms of α-factor were detected, including pro-α-factor (pαf) that includes a range of high molecular weight intermediates containing the proregion and various degrees Golgi-derived glycosylation (∼ 90– 150 kDa), unglycosylated core-α-factor (unglyc) (∼20 kDa), ER core-glycosylated α-factor (core) (∼26 kDa), low molecular weight intermediates in α-factor processing (interαf) that most likely represent the tetrapeptide in various states of processing with the proregion removed (∼6–8 kDa), and the mature α-factor (mαf), the 13-amino acid terminally processed secreted form (∼2 kDa).
Similar articles
- Identification and characterization of Saccharomyces cerevisiae mutants defective in fluid-phase endocytosis.
Wiederkehr A, Meier KD, Riezman H. Wiederkehr A, et al. Yeast. 2001 Jun;18(8):759-73. doi: 10.1002/yea.726. Yeast. 2001. PMID: 11378903 - Compartment acidification is required for efficient sorting of proteins to the vacuole in Saccharomyces cerevisiae.
Klionsky DJ, Nelson H, Nelson N. Klionsky DJ, et al. J Biol Chem. 1992 Feb 15;267(5):3416-22. J Biol Chem. 1992. PMID: 1531340 - Vacuolar sorting. Tracking down an elusive receptor.
Chapman RE. Chapman RE. Curr Biol. 1994 Nov 1;4(11):1019-22. doi: 10.1016/s0960-9822(00)00231-1. Curr Biol. 1994. PMID: 7874484 Review. - Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins.
Stack JH, Horazdovsky B, Emr SD. Stack JH, et al. Annu Rev Cell Dev Biol. 1995;11:1-33. doi: 10.1146/annurev.cb.11.110195.000245. Annu Rev Cell Dev Biol. 1995. PMID: 8689553 Review.
Cited by
- Identification of yeast genes that confer resistance to chitosan oligosaccharide (COS) using chemogenomics.
Jaime MD, Lopez-Llorca LV, Conesa A, Lee AY, Proctor M, Heisler LE, Gebbia M, Giaever G, Westwood JT, Nislow C. Jaime MD, et al. BMC Genomics. 2012 Jun 22;13:267. doi: 10.1186/1471-2164-13-267. BMC Genomics. 2012. PMID: 22727066 Free PMC article. - Broad network-based predictability of Saccharomyces cerevisiae gene loss-of-function phenotypes.
McGary KL, Lee I, Marcotte EM. McGary KL, et al. Genome Biol. 2007;8(12):R258. doi: 10.1186/gb-2007-8-12-r258. Genome Biol. 2007. PMID: 18053250 Free PMC article. - Multiple signals converge on a differentiation MAPK pathway.
Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. Chavel CA, et al. PLoS Genet. 2010 Mar 19;6(3):e1000883. doi: 10.1371/journal.pgen.1000883. PLoS Genet. 2010. PMID: 20333241 Free PMC article. - Genome-wide identification and analysis of membrane-bound O-acyltransferase (MBOAT) gene family in plants.
Wang P, Wang Z, Dou Y, Zhang X, Wang M, Tian X. Wang P, et al. Planta. 2013 Nov;238(5):907-22. doi: 10.1007/s00425-013-1939-4. Epub 2013 Aug 9. Planta. 2013. PMID: 23928653 - The MAP kinase Slt2 is involved in vacuolar function and actin remodeling in Saccharomyces cerevisiae mutants affected by endogenous oxidative stress.
Pujol-Carrion N, Petkova MI, Serrano L, de la Torre-Ruiz MA. Pujol-Carrion N, et al. Appl Environ Microbiol. 2013 Oct;79(20):6459-71. doi: 10.1128/AEM.01692-13. Epub 2013 Aug 16. Appl Environ Microbiol. 2013. PMID: 23956390 Free PMC article.
References
- Bell AW, et al. Proteomics characterization of abundant Golgi membrane proteins. J Biol Chem. 2001;276:5152–65. - PubMed
- Bonifacino JS, Dell'Angelica EC. Immunoprecipitation. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada K, editors. Current Protocols in Cell Biology. New York: John Wiley & Sons; 1998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases