Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration - PubMed (original) (raw)
Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration
Howard A Ross et al. J Virol. 2002 Nov.
Abstract
Using likelihood-based evolutionary methods, we demonstrate that the broad genetic diversity of human immunodeficiency virus type 1 (HIV-1) in an infected individual is a consequence of site-specific positive selection for diversity, a likely consequence of immune recognition. In particular, the extent of positive selection appears to be a good predictor of disease duration. Positively selected sites along HIV-1 partial env sequences are numerous but not distributed uniformly. In a sample of eight patients studied longitudinally, the proportion of sites per sample under positive selection was a statistically significant predictor of disease duration. Among long-term progressors, positive selection persisted at sites over time and appears to be associated with helper T-cell epitopes. In contrast, sites under positive selection shifted from one longitudinal sample to the next in short-term progressors. Our study is consistent with the hypothesis that a broad and persistent immunologic response is associated with a slower rate of disease progression. In contrast, narrow, shifting immune responses characterize short-term progressors.
Figures
FIG. 1.
The location of sites classified as experiencing positive selection in the C2-V5 region of the HIV-1 env gene. Individual sites are color coded when ω is >1 in at least one sample. The proportion of sites with ω > 1 is indicated as red (<0.01), yellow (0.01 to 0.03), green (0.03 to 0.07), light blue (0.07 to 0.15), dark blue (>0.15), or grey (no sequence). Sequences are arranged vertically, with one column per sample. Subjects are arranged from left to right in decreasing order of longevity or, for subjects with the same longevity, in decreasing proportion of positively selected sites. Within subjects, samples are arranged from left to right by time of collection. Column headings are color coded to indicate the observed proportion of positively selected sites within the sample or subject. Alignment with external reference sequences and gap balancing (32) created gaps.
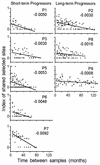
FIG. 2.
Persistence of positive selection in the two groups of subjects. The proportion of sites under positive selection in both of two samples, in all pair-wise comparisons, is plotted against the time interval between samples. The results for each subject are plotted separately. The plots are labeled with the patient identifier and the slope of the line of best fit.
Similar articles
- Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes.
Choisy M, Woelk CH, Guégan JF, Robertson DL. Choisy M, et al. J Virol. 2004 Feb;78(4):1962-70. doi: 10.1128/jvi.78.4.1962-1970.2004. J Virol. 2004. PMID: 14747561 Free PMC article. - Lack of temporal structure in the short term HIV-1 evolution within asymptomatic naïve patients.
Bello G, Casado C, García S, Rodríguez C, del Romero J, Carvajal-Rodriguez A, Posada D, López-Galíndez C. Bello G, et al. Virology. 2007 Jun 5;362(2):294-303. doi: 10.1016/j.virol.2006.11.039. Epub 2007 Jan 31. Virology. 2007. PMID: 17275055 - Molecular clock of HIV-1 envelope genes under early immune selection.
Park SY, Love TM, Perelson AS, Mack WJ, Lee HY. Park SY, et al. Retrovirology. 2016 Jun 1;13(1):38. doi: 10.1186/s12977-016-0269-6. Retrovirology. 2016. PMID: 27246201 Free PMC article. - Human immunodeficiency virus type 1 molecular evolution and the measure of selection.
Rodrigo AG, Mullins JI. Rodrigo AG, et al. AIDS Res Hum Retroviruses. 1996 Dec 10;12(18):1681-5. doi: 10.1089/aid.1996.12.1681. AIDS Res Hum Retroviruses. 1996. PMID: 8959243 - Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression.
Poropatich K, Sullivan DJ Jr. Poropatich K, et al. J Gen Virol. 2011 Feb;92(Pt 2):247-68. doi: 10.1099/vir.0.027102-0. Epub 2010 Nov 24. J Gen Virol. 2011. PMID: 21106806 Review.
Cited by
- HIV-1 diversity in viral reservoirs obtained from circulating T-cell subsets during early ART and beyond.
Zhang Y, Otte F, Stoeckle M, Thielen A, Däumer M, Kaiser R, Kusejko K, Metzner KJ, Klimkait T; and the Swiss HIV Cohort Study. Zhang Y, et al. PLoS Pathog. 2024 Sep 18;20(9):e1012526. doi: 10.1371/journal.ppat.1012526. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39292732 Free PMC article. - Machine Learning Prediction and Phyloanatomic Modeling of Viral Neuroadaptive Signatures in the Macaque Model of HIV-Mediated Neuropathology.
Ramirez-Mata AS, Ostrov D, Salemi M, Marini S, Magalis BR. Ramirez-Mata AS, et al. Microbiol Spectr. 2023 Feb 27;11(2):e0308622. doi: 10.1128/spectrum.03086-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36847516 Free PMC article. - Longitudinal within-host evolution of HIV Nef-mediated CD4, HLA and SERINC5 downregulation activity: a case study.
Sudderuddin H, Kinloch NN, Jin SW, Miller RL, Jones BR, Brumme CJ, Joy JB, Brockman MA, Brumme ZL. Sudderuddin H, et al. Retrovirology. 2020 Jan 9;17(1):3. doi: 10.1186/s12977-019-0510-1. Retrovirology. 2020. PMID: 31918727 Free PMC article. - Low Postseroconversion CD4+ T-cell Level Is Associated with Faster Disease Progression and Higher Viral Evolutionary Rate in HIV-2 Infection.
Palm AA, Lemey P, Jansson M, Månsson F, Kvist A, Szojka Z, Biague A, da Silva ZJ, Rowland-Jones SL, Norrgren H, Esbjörnsson J, Medstrand P. Palm AA, et al. mBio. 2019 Jan 8;10(1):e01245-18. doi: 10.1128/mBio.01245-18. mBio. 2019. PMID: 30622192 Free PMC article. - HIV Progression Depends on Codon and Amino Acid Usage Profile of Envelope Protein and Associated Host-Genetic Influence.
Roy A, Banerjee R, Basak S. Roy A, et al. Front Microbiol. 2017 Jun 15;8:1083. doi: 10.3389/fmicb.2017.01083. eCollection 2017. Front Microbiol. 2017. PMID: 28663742 Free PMC article.
References
- Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. - PubMed
- Bonhoeffer, S., E. C. Holmes, and M. A. Nowak. 1995. Causes of HIV diversity. Nature 376:125. - PubMed
- Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. - PubMed
- Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major coreceptor for primary isolates of HIV-1. Nature 381:661-666. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical