Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins - PubMed (original) (raw)
Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins
D L Lindstrom et al. Mol Cell Biol. 2003 Feb.
Abstract
During transcription elongation, eukaryotic RNA polymerase II (Pol II) must contend with the barrier presented by nucleosomes. The conserved Spt4-Spt5 complex has been proposed to regulate elongation through nucleosomes by Pol II. To help define the mechanism of Spt5 function, we have characterized proteins that coimmunopurify with Spt5. Among these are the general elongation factors TFIIF and TFIIS as well as Spt6 and FACT, factors thought to regulate elongation through nucleosomes. Spt5 also coimmunopurified with the mRNA capping enzyme and cap methyltransferase, and spt4 and spt5 mutations displayed genetic interactions with mutations in capping enzyme genes. Additionally, we found that spt4 and spt5 mutations lead to accumulation of unspliced pre-mRNA. Spt5 also copurified with several previously unstudied proteins; we demonstrate that one of these is encoded by a new member of the SPT gene family. Finally, by immunoprecipitating these factors we found evidence that Spt5 participates in at least three Pol II complexes. These observations provide new evidence of roles for Spt4-Spt5 in pre-mRNA processing and transcription elongation.
Figures
FIG. 1.
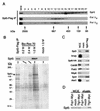
Identification of Spt5-associated proteins. (A) Anti-Flag immunoprecipitates from extracts of an Spt5-Flag strain (GHY617) were size fractionated by gel filtration, and fractions were analyzed by blotting with antibodies specific for Spt5, the hypophosphorylated form of Rpb1 (Pol IIA), and the hyperphosphorylated form of Rpb1 (Pol IIO). Control experiments demonstrated that coprecipitation of both Pol II isoforms with Spt5-Flag was specific (reference and data not shown). (B) Spt5-Flag purification. Extracts of an Spt5-Flag strain and an untagged strain (mock) were incubated with anti-Flag M2 agarose beads. Spt5-Flag complexes were competitively eluted from the beads with Flag peptide and further fractionated on a Bio-Rex 70 column. Fractions were analyzed by silver staining and blotting for Spt5. Upper panel, anti-Spt5 Western blot. Lower panel, silver-stained gel. (C) Anti-Flag immunoprecipitations from extracts of Spt5-Flag and untagged (mock) strains. Proteins were eluted with Flag peptide, separated by SDS-PAGE, and blotted with the indicated antibodies. WCE, 30 μg of whole-cell extract of Spt5-Flag strain. Note that in this panel the relative proportion of whole-cell extract to eluate is five times higher than in other immunoprecipitations. (D) Anti-Myc immunoprecipitations from extracts of strains carrying Fcp1-Myc or Tfg1-Myc and a strain lacking the Myc tag (mock). Proteins were eluted in 1.0 M potassium acetate, fractionated by SDS-PAGE, and blotted for Spt5. Strains: Spt5-Flag, GHY617; Fcp1-Myc, GHY1207; Tfg1-Myc, GHY1208; mock in panels A to C, GHY611; mock in panel D, GHY617.
FIG. 2.
Immunoprecipitation of Spt4, Spt6, Cdc68, and Pob3. (A) Anti-Flag immunoprecipitation from lysates of an Spt4-Flag strain and an untagged strain (mock). Bound proteins were eluted from the beads in 1.0 M potassium acetate, fractionated by SDS-PAGE, and Western blotted for the indicated proteins. Strains: mock, GHY1320; Spt4-Flag, GHY1324. (B) Anti-HA immunoprecipitations from lysates of an HA-Spt6 strain and an untagged strain were performed as for panel A. Strains: mock, GHY611; HA-Spt6, GHY605. WCE, 30 μg of whole-cell extract. (C) Anti-HA immunoprecipitations from lysates of a HA-Spt6 and an untagged strain were performed as for panel A. Strains: mock, GHY1325; HA-Spt6, GHY1324. WCE, 60 μg of whole-cell extract. (D) Anti-Myc immunoprecipitations from extracts of Cdc68-Myc and untagged (mock) strains were performed as for panel A. Top panel, 60 μg of whole-cell extract and 10% of the anti-Myc beads from the immunoprecipitations were separated by SDS-PAGE and blotted for the presence of Cdc68-Myc and Pob3. In contrast to the other proteins analyzed in panels D and E, Cdc68-Pob3 complexes are stable in 1.0 M potassium acetate and therefore remain bound to the beads (data not shown). Bottom panel, 60 μg of whole-cell extract and 1 M potassium acetate eluates from the anti-Myc beads were separated by SDS-PAGE and blotted for the presence of HA-Spt6 and Abd1. The asterisk indicates a weak cross-reactivity to immunoglobulin G migrating near the expected position of Abd1 in the gel. Strains: Cdc68-Myc, OY240; mock, GHY605. (E) Anti-Myc immunoprecipitations from extracts of Pob3-Myc, Cdc68-Myc, and untagged (mock) strains were performed as for panel A. Top panel, 30 μg of whole-cell lysates and 1 M potassium acetate eluates were separated by SDS-PAGE and probed for the indicated proteins. Bottom panel, 30 μg of whole-cell extract and 10% of the anti-Myc beads from the immunoprecipitations were separated by SDS-PAGE and blotted with an anti-Myc antibody for the presence of Cdc68-Myc and Pob3-Myc. Strains used: Pob3-Myc, DY6460; Cdc68-Myc, DY6529; untagged, DK186.
FIG. 3.
Spt5 but not Pob3 associates with capping enzyme and cap methyltransferase. Anti-HA immunoprecipitations from extracts of HA-Ceg1, HA-Abd1, and untagged (mock) strains are shown. Bound proteins were eluted with 1.0 M potassium acetate, separated by SDS-PAGE, and blotted for Spt5 and Pob3. WCE, 30 μg of whole-cell extract. Eluate, 1.0 M potassium acetate elutions. Strains: HA-Ceg1, YSB244/pSB996; HA-Abd1, YSB427/pSB995; untagged, YSB244/pRS316-CEG1.
FIG. 4.
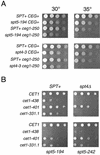
SPT4 and SPT5 display genetic interactions with capping enzyme. (A) Strains with the indicated genotypes were assayed by serial dilution onto YPD plates, incubated for 2 days at the indicated temperatures, and photographed. Note that the restrictive temperature for the _ceg1_-250 single mutant is 37°C (19). Strains: wild type, FY120; _spt5_-194, GHY594; _spt4_-3, GHY96; _ceg1_-250, OY163; _spt5_-_194 ceg1_-250, OY215; _spt4_-_3 ceg1_-250, OY167. (B) spt cet1Δ mutants carrying a URA3 CET1 plasmid were transformed with a series of HIS3 cet1 plasmids as indicated. Transformants were grown to saturation in SC-histidine medium and assayed by serial dilution on 5-fluoroorotic acid plates, which allows growth only of cells that have lost the URA3 CET1 plasmid. Plates were incubated for 2 days at 30°C and photographed. Strains: cet1Δ1::TRP, OY227; spt4Δ cet1Δ1::TRP1, OY204; _spt5_-194 cet1Δ1::TRP1, OY207; _spt5_-242 cet1Δ1::TRP1, OY205.
FIG. 5.
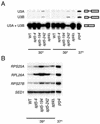
Splicing defects in spt4 and spt5 mutants. RNA was prepared from strains with the indicated genotypes either grown at 30°C or grown at 30°C and then shifted to 39°C for 45 min prior to harvest. The prp4 strain was grown at 30°C and then shifted to 37°C for 1 h prior to harvest. (A) Primer extension analysis of the U3A and U3B snRNAs. Note that mature U3A and U3B snRNAs are indistinguishable in this assay. In contrast, the unspliced U3A and U3B pre-snRNAs give products that differ by 27 nucleotides (4). (B) RT-PCR analysis for unspliced forms of the RPS25A, RPL26A, and RPS27B genes. Unspliced pre-mRNAs were specifically amplified by using one primer chosen from intron sequences and another chosen from the second exon. As a loading control, SED1, which does not contain an intron, was also analyzed by RT-PCR. Control reactions confirmed that the PCRs were performed in the linear range and that the PCR products were dependent upon reverse transcriptase (data not shown). Strains: wild type (WT), FY120; _spt5_-4, OY43; _spt5_-194, GHY379; _spt5_-242, GHY92; spt4Δ, GHY524; prp4, SRY4-1a.
FIG. 6.
IWS1/YPR133C encodes a conserved Spt5-associated protein required for normal transcription. (A) Alignment of Iws1 and homologs. The regions of highest identity between the proteins are marked by shading, and the percent amino acid identity is noted. The N-terminal SR repeats in the Drosophila homolog are also noted by shading. The accession numbers for the human and Drosophila homologs are BAA91402 and AAF48587, respectively. Alignments and percent amino acid identities were determined by BLAST searches (1). (B) Anti-Myc immunoprecipitations were performed from extracts of an Iws1-Myc strain and an untagged (mock) strain. Bound proteins were eluted with 1.0 M potassium acetate, separated by SDS-PAGE, and blotted for Spt5. Strains: Iws1-Myc, GHY1300; mock, GHY1280. WCE, 30 μg of whole-cell extract of GHY1300. (C) iws1 mutants display an Spt− phenotype. Cells were replica plated to the indicated media and grown for 2 days at 30°C. Strains: Spt+, GHY611; _iws1_-13, GHY1202; _spt5_-4, GHY1073. (D) The Spt− phenotypes of _iws1_-7 and _iws1_-13 cells are due to altered transcription. Top panel, Northern blot analysis of HIS4 and _his4_-912δ RNA derived from strains with the indicated genotypes. RNA from a _spt5_-_194 his4_-912δ strain was included for comparison to the _iws1 his4_-912δ strains. Bottom panel, as a loading control, the blot was stripped and then rehybridized with a TUB2 probe. Strains: lane 1, FY602; lane 2, FY119; lane 3, GHY13; lane 4, GHY1199; lane 5, GHY1200.
FIG. 7.
Model of Spt5 complexes. This model is based on coimmunoprecipitation and purification data presented in Fig. 1, 2, 3, and 6 and on previous studies demonstrating that Abd1 and the capping enzyme interact specifically with Pol IIO (11, 36, 68). The presence of Spt4, Spt6, and Iws1 in the same complex as the capping enzyme remains to be tested directly. For this reason, the capping enzyme complex in panel B is indicated with a dashed line.
Similar articles
- Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae.
Hartzog GA, Wada T, Handa H, Winston F. Hartzog GA, et al. Genes Dev. 1998 Feb 1;12(3):357-69. doi: 10.1101/gad.12.3.357. Genes Dev. 1998. PMID: 9450930 Free PMC article. - RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach.
Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. Krogan NJ, et al. Mol Cell Biol. 2002 Oct;22(20):6979-92. doi: 10.1128/MCB.22.20.6979-6992.2002. Mol Cell Biol. 2002. PMID: 12242279 Free PMC article. - Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest.
Crickard JB, Fu J, Reese JC. Crickard JB, et al. J Biol Chem. 2016 May 6;291(19):9853-70. doi: 10.1074/jbc.M116.716001. Epub 2016 Mar 4. J Biol Chem. 2016. PMID: 26945063 Free PMC article. - The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation.
Hartzog GA, Fu J. Hartzog GA, et al. Biochim Biophys Acta. 2013 Jan;1829(1):105-15. doi: 10.1016/j.bbagrm.2012.08.007. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982195 Free PMC article. Review. - The pleiotropic roles of SPT5 in transcription.
Song A, Chen FX. Song A, et al. Transcription. 2022 Feb-Jun;13(1-3):53-69. doi: 10.1080/21541264.2022.2103366. Epub 2022 Jul 25. Transcription. 2022. PMID: 35876486 Free PMC article. Review.
Cited by
- Network compression as a quality measure for protein interaction networks.
Royer L, Reimann M, Stewart AF, Schroeder M. Royer L, et al. PLoS One. 2012;7(6):e35729. doi: 10.1371/journal.pone.0035729. Epub 2012 Jun 18. PLoS One. 2012. PMID: 22719828 Free PMC article. - The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex.
Mayekar MK, Gardner RG, Arndt KM. Mayekar MK, et al. Mol Cell Biol. 2013 Aug;33(16):3259-73. doi: 10.1128/MCB.00270-13. Epub 2013 Jun 17. Mol Cell Biol. 2013. PMID: 23775116 Free PMC article. - Comprehensive analysis of m7G modification patterns based on potential m7G regulators and tumor microenvironment infiltration characterization in lung adenocarcinoma.
Ma S, Zhu J, Wang M, Zhu J, Wang W, Xiong Y, Jiang R, Liu L, Jiang T. Ma S, et al. Front Genet. 2022 Sep 29;13:996950. doi: 10.3389/fgene.2022.996950. eCollection 2022. Front Genet. 2022. PMID: 36246663 Free PMC article. - Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment.
Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Eissenberg JC, et al. Mol Genet Genomics. 2007 Feb;277(2):101-14. doi: 10.1007/s00438-006-0164-2. Epub 2006 Sep 26. Mol Genet Genomics. 2007. PMID: 17001490 - Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants.
Jimeno S, Luna R, García-Rubio M, Aguilera A. Jimeno S, et al. Mol Cell Biol. 2006 Jun;26(12):4387-98. doi: 10.1128/MCB.00234-06. Mol Cell Biol. 2006. PMID: 16738307 Free PMC article.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Archambault, J., R. S. Chambers, M. S. Kobor, Y. Ho, M. Cartier, D. Bolotin, B. Andrews, C. M. Kane, and J. Greenblatt. 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:14300-14305. - PMC - PubMed
- Ares, M., Jr., and A. H. Igel. 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 4:2132-2145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous