Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells - PubMed (original) (raw)
Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells
Philippe Giegé et al. Plant Cell. 2003 Sep.
Abstract
Mitochondria fulfill a wide range of metabolic functions in addition to the synthesis of ATP and contain a diverse array of proteins to perform these functions. Here, we present the unexpected discovery of the presence of the enzymes of glycolysis in a mitochondrial fraction of Arabidopsis cells. Proteomic analyses of this mitochondrial fraction revealed the presence of 7 of the 10 enzymes that constitute the glycolytic pathway. Four of these enzymes (glyceraldehyde-3-P dehydrogenase, aldolase, phosphoglycerate mutase, and enolase) were also identified in an intermembrane space/outer mitochondrial membrane fraction. Enzyme activity assays confirmed that the entire glycolytic pathway was present in preparations of isolated Arabidopsis mitochondria, and the sensitivity of these activities to protease treatments indicated that the glycolytic enzymes are present on the outside of the mitochondrion. The association of glycolytic enzymes with mitochondria was confirmed in vivo by the expression of enolase- and aldolase-yellow fluorescent protein fusions in Arabidopsis protoplasts. The yellow fluorescent protein fluorescence signal showed that these two fusion proteins are present throughout the cytosol but are also concentrated in punctate regions that colocalized with the mitochondrion-specific probe Mitotracker Red. Furthermore, when supplied with appropriate cofactors, isolated, intact mitochondria were capable of the metabolism of (13)C-glucose to (13)C-labeled intermediates of the trichloroacetic acid cycle, suggesting that the complete glycolytic sequence is present and active in this subcellular fraction. On the basis of these data, we propose that the entire glycolytic pathway is associated with plant mitochondria by attachment to the cytosolic face of the outer mitochondrial membrane and that this microcompartmentation of glycolysis allows pyruvate to be provided directly to the mitochondrion, where it is used as a respiratory substrate.
Figures
Figure 1.
Proteins of the IMS and OMM Separated by Two-Dimensional Gel Electrophoresis. An IMS/OMM fraction was prepared from isolated Arabidopsis mitochondria by osmotic shock and centrifugation (for details, see Methods). Proteins were separated by isoelectric focusing on 3 to 10 nonlinear immobilized pH gradients for the first dimension and by SDS-PAGE for the second dimension. For clarity, only the relevant section of the gel is shown. The numbered spots are those that have been identified by MS/MS (see Table 2).
Figure 2.
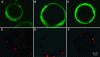
Localization of Aldolase and Enolase in Arabidopsis Protoplasts. Arabidopsis protoplasts were transformed with constructs for the expression of an aldolase-YFP fusion, an enolase-YFP fusion, and YFP alone (for more details, see Methods). YFP fluorescence was visualized using a confocal microscope and compared with the distribution of the mitochondrion-specific probe Mitotracker Red (Molecular Probes). (A) to (C) YFP fluorescence. (D) to (F) Mitotracker Red fluorescence. (A) and (D) show a protoplast expressing aldolase::YFP, (B) and (E) show a protoplast expressing enolase::YFP, and (C) and (F) show a protoplast expressing YFP alone as a control.
Figure 3.
Submitochondrial Localization of Aldolase Is Revealed by a Protease Protection Assay. The protease thermolysin was added to isolated mitochondria in the presence or absence of Triton X-100 to rupture the mitochondrial membranes. The reaction was terminated by the addition of EGTA, and 50 μg of the protein sample was fractionated by SDS-PAGE before being transferred to a nitrocellulose membrane. The membrane was probed with antisera against aldolase and fumarase.
Similar articles
- A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts.
Zhang Y, Sampathkumar A, Kerber SM, Swart C, Hille C, Seerangan K, Graf A, Sweetlove L, Fernie AR. Zhang Y, et al. Nat Commun. 2020 Sep 9;11(1):4509. doi: 10.1038/s41467-020-18234-w. Nat Commun. 2020. PMID: 32908151 Free PMC article. - Glycolytic enzyme interactions with tubulin and microtubules.
Walsh JL, Keith TJ, Knull HR. Walsh JL, et al. Biochim Biophys Acta. 1989 Nov 9;999(1):64-70. doi: 10.1016/0167-4838(89)90031-9. Biochim Biophys Acta. 1989. PMID: 2553125 - Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling.
Graham JW, Williams TC, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ. Graham JW, et al. Plant Cell. 2007 Nov;19(11):3723-38. doi: 10.1105/tpc.107.053371. Epub 2007 Nov 2. Plant Cell. 2007. PMID: 17981998 Free PMC article. - Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells.
Fukushi A, Kim HD, Chang YC, Kim CH. Fukushi A, et al. Int J Mol Sci. 2022 Sep 2;23(17):10037. doi: 10.3390/ijms231710037. Int J Mol Sci. 2022. PMID: 36077431 Free PMC article. Review. - X-ray studies of glycolytic enzymes.
Blake CC. Blake CC. Essays Biochem. 1975;11:37-79. Essays Biochem. 1975. PMID: 174910 Review. No abstract available.
Cited by
- The OsGAPC3 mutation significantly affects grain quality traits and improves the nutritional quality of rice.
Peng B, Liu Y, Sun X, Zhao Q, Qiu J, Tian X, Peng J, Zhang Z, Wang Y, Huang Y, Pang R, Zhou W, Qi Y, Sun Y, Wang Q, He Y. Peng B, et al. Front Plant Sci. 2024 Oct 3;15:1470316. doi: 10.3389/fpls.2024.1470316. eCollection 2024. Front Plant Sci. 2024. PMID: 39421143 Free PMC article. - Elucidating the Role of Human ALAS2 C-terminal Mutations Resulting in Loss of Function and Disease.
Taylor JL, Ayres-Galhardo PH, Brown BL. Taylor JL, et al. Biochemistry. 2024 Jul 2;63(13):1636-1646. doi: 10.1021/acs.biochem.4c00066. Epub 2024 Jun 18. Biochemistry. 2024. PMID: 38888931 Free PMC article. - Plastid Transient and Stable Interactions with Other Cell Compartments.
Mueller-Schuessele SJ, Leterme S, Michaud M. Mueller-Schuessele SJ, et al. Methods Mol Biol. 2024;2776:107-134. doi: 10.1007/978-1-0716-3726-5_6. Methods Mol Biol. 2024. PMID: 38502500 - Unique Features of the m6A Methylome and Its Response to Salt Stress in the Roots of Sugar Beet (Beta vulgaris).
Li J, Pang Q, Yan X. Li J, et al. Int J Mol Sci. 2023 Jul 19;24(14):11659. doi: 10.3390/ijms241411659. Int J Mol Sci. 2023. PMID: 37511417 Free PMC article. - Mitochondrial Proteome Changes in Rett Syndrome.
Golubiani G, van Agen L, Tsverava L, Solomonia R, Müller M. Golubiani G, et al. Biology (Basel). 2023 Jul 3;12(7):956. doi: 10.3390/biology12070956. Biology (Basel). 2023. PMID: 37508386 Free PMC article.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
- Azama, K., Abe, S., Sugimoto, H., and Davies, E. (2003). Lysine-containing proteins in maize endosperm: A major contribution from cytoskeleton-associated carbohydrate-metabolizing enzymes. Planta, in press. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases