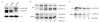Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly - PubMed (original) (raw)
Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly
Marion Fehlker et al. EMBO Rep. 2003 Oct.
Abstract
Proteasomes are multisubunit proteases that are responsible for regulated proteolysis. The degradation of the proteasomal maturation factor, named Ump1 in yeast, completes the autocatalytic processing of inactive precursor complexes into the proteolytically active core particle (CP) of the proteasome. We have identified Blm3, a conserved nuclear protein, as a new component of Ump1-associated precursor complexes. A lack of Blm3 resulted in an increased rate of precursor processing and an accelerated turnover of Ump1, which suggests that Blm3 prevents premature activation of proteasomal CPs. On the basis of biochemical fractionation experiments combined with in vivo localization studies, we propose that Blm3 joins nascent CPs inside the nucleus to coordinate late stages of proteasome assembly in yeast.
Figures
Figure 1
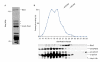
Identification of Blm3 as part of Ump1-associated precursor complexes. (A) Coomassie-blue-stained SDS–polyacrylamide gel of Ump1-associated precursor complexes that were isolated by IgG affinity-chromatography from nuclear/endoplasmic reticulum extracts from yeast wild-type cells expressing protein A (ProA)-tagged Ump1 instead of the endogenous protein (Lehmann et al., 2002). Blm3 and Ump1–ProA are indicated. Proteins indicated by asterisks are heat shock proteins. (B) Extracts from wild-type cells expressing haemagglutinin (HA)-tagged β5 and Ump1 instead of the endogenous proteins (wild-type strain JD133; Ramos et al., 1998) were fractionated on Superose 6 (Amersham). Fractions were assayed for peptide-cleavage activity against Cbz-Leu-Leu-Glu-β-naphthylamide (upper panel). Protein samples of each fraction were run on SDS–polyacrylamide gels, blotted and probed for Blm3, the regulatory particle subunit Cim3/Rpt6, the core particle subunit β5–HA and Ump1–HA (lower panel). Unprocessed and matured β5 subunits are abbreviated as pro-β5 and m-β5, respectively. A degradation band of m-β5 is indicated by an asterisk. The Superose 6 column was calibrated using the standards thyroglobulin (670 kDa) and ferritin (440 kDa).
Figure 2
Blm3 delays core particle maturation. (A) Wild-type (WT),Δblm3 and Δump1 cells were grown to logarithmic phase (optical density of 1 at 600 nm), harvested, boiled immediately in sample buffer and subjected to SDS–polyacrylamide gel electrophoresis followed by immunoblotting. Proteins were probed for haemagglutinin (HA)-tagged β5 and α4 subunits, the latter of which was used as a loading control. Unprocessed (pro-) β5, matured (m-) β5 and α4 are indicated. (B) Pulse-chase analysis, comparing the rates of pro-β5 processing and Ump1 degradation in wild-type and blm3Δ cells. Cells were pulse-labelled with 35S-Met/Cys for 5 min and chased for the lengths of time indicated. HA-tagged β5 and Ump1 were precipitated with anti-HA antibodies. Pro-β5, m-β5 and Ump1 are indicated. Each pulse-chase analysis comparing blm3Δ and wild-type cells was performed three times in parallel.
Figure 3
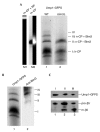
Blm3 is associated with the nascent core particle but not with the half-assembled core particle. (A) Green-fluorescent-protein–streptactin (GFPS)-tagged precursor complexes, which were isolated by streptactin affinity chromatography from wild-type (WT; lane 1) and blm3Δ (lane 2) cells expressing Ump1–GFPS instead of the endogenous protein, respectively, were separated by native polyacrylamide gel electrophoresis. The GFP-labelled precursor complexes were visualized by scanning the gel with a phosphofluoroimager. Bands were numbered (I–IV) and assigned to the half-assembled core particle (h-CP), or to the nascent CP (n-CP) with (+) or without (−) Blm3. Species I was assigned to the h-CP, as this species represents the main fraction of precursor complexes (Chen & Hochstrasser, 1996; Ramos et al., 1998; Lehmann et al., 2002). The n-CP was found to behave like the mature CP (m-CP) in native gels. Thus, the m-CP (Lehmann et al., 2002) was run as a size marker (lane M2; the left half shows phosphofluoroimaging of the GFPS-tagged m-CP; the right half shows the chromogenic peptide-cleavage activity of the GFPS-tagged m-CP. As another control, the migration of the m-CP in association with the regulatory particle (RP) is shown (lane M1). (B) Native gels of Ump1-associated precursor complexes were scanned for Ump1–GFPS (lane 1), blotted, and probed for Blm3 (lane 2). Bands were assigned as above. Band IV probably represents the n-CP capped by two Blm3 molecules, but could not be characterized further due to limiting amounts of protein. (C) The wild-type precursor complex species (bands I–III) were excised from the native gel, subjected to SDS–PAGE, blotted, and probed for Ump1 and β5 (lanes 1–3 are derived from bands I–III, respectively). Ump1–GFPS, pro-β5 and mature β5 (m-β5) are indicated. Subunits of the RP were not detected in any of our preparations of Ump1-associated precursor complexes (data not shown).
Figure 4
Blm3 is nuclear. (A) Western blot analysis of cells in which Blm3 is chromosomally replaced by a green fluorescent protein (GFP)-tagged version (lane 1) compared with mock-transfected (mock) cells (lane 2). Anti-GFP antibodies were used. (B) Direct fluorescence microscopy was used (Enenkel et al., 1998) to monitor GFP-tagged Blm3 in living cells (left panel, DAPI (4′,6-diamidino-2-phenylindole) staining of yeast nuclei, as visualized by the ultraviolet light filter superimposed with Nomarski optics; right panel, GFP filter superimposed with Nomarski optics). (C) Spheroplast lysates were fractionated into a cytosolic (C) and a nuclear (N) fraction, separated by SDS–polyacrylamide gel electrophoresis, blotted, and probed for Blm3, nuclear (Nups) and cytoplasmic (PFK) controls (Enenkel et al., 1998).
Similar articles
- 20S proteasome biogenesis.
Krüger E, Kloetzel PM, Enenkel C. Krüger E, et al. Biochimie. 2001 Mar-Apr;83(3-4):289-93. doi: 10.1016/s0300-9084(01)01241-x. Biochimie. 2001. PMID: 11295488 Review. - 20 S proteasomes are imported as precursor complexes into the nucleus of yeast.
Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C. Lehmann A, et al. J Mol Biol. 2002 Mar 29;317(3):401-13. doi: 10.1006/jmbi.2002.5443. J Mol Biol. 2002. PMID: 11922673 - The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation.
Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. Marques AJ, et al. J Biol Chem. 2007 Nov 30;282(48):34869-76. doi: 10.1074/jbc.M705836200. Epub 2007 Oct 2. J Biol Chem. 2007. PMID: 17911101 - Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly.
Chen P, Hochstrasser M. Chen P, et al. Cell. 1996 Sep 20;86(6):961-72. doi: 10.1016/s0092-8674(00)80171-3. Cell. 1996. PMID: 8808631 - Proteasome assembly: biting the hand.
Maurizi MR. Maurizi MR. Curr Biol. 1998 Jun 18;8(13):R453-6. doi: 10.1016/s0960-9822(98)70291-x. Curr Biol. 1998. PMID: 9651672 Review.
Cited by
- Structural roles of Ump1 and β-subunit propeptides in proteasome biogenesis.
Mark E, Ramos PC, Kayser F, Höckendorff J, Dohmen RJ, Wendler P. Mark E, et al. Life Sci Alliance. 2024 Sep 11;7(11):e202402865. doi: 10.26508/lsa.202402865. Print 2024 Nov. Life Sci Alliance. 2024. PMID: 39260885 Free PMC article. - Assembly of the 20S proteasome.
Kunjappu MJ, Hochstrasser M. Kunjappu MJ, et al. Biochim Biophys Acta. 2014 Jan;1843(1):2-12. doi: 10.1016/j.bbamcr.2013.03.008. Epub 2013 Mar 16. Biochim Biophys Acta. 2014. PMID: 23507199 Free PMC article. Review. - Role of the Ubiquitin-Proteasome Systems in the Biology and Virulence of Protozoan Parasites.
Muñoz C, San Francisco J, Gutiérrez B, González J. Muñoz C, et al. Biomed Res Int. 2015;2015:141526. doi: 10.1155/2015/141526. Epub 2015 May 19. Biomed Res Int. 2015. PMID: 26090380 Free PMC article. Review. - A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding.
Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. Kusmierczyk AR, et al. Nat Struct Mol Biol. 2011 May;18(5):622-9. doi: 10.1038/nsmb.2027. Epub 2011 Apr 17. Nat Struct Mol Biol. 2011. PMID: 21499243 Free PMC article. - Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism.
Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M. Dange T, et al. J Biol Chem. 2011 Dec 16;286(50):42830-9. doi: 10.1074/jbc.M111.300178. Epub 2011 Oct 24. J Biol Chem. 2011. PMID: 22025621 Free PMC article.
References
- Chen P. & Hochstrasser M. ( 1996) Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell, 86, 961–972. - PubMed
- Evans Febres D., Pramanik A., Caton M., Doherty K., McKoy J., Garcia E., Alejo W. & Moore C.W. ( 2001) The novel BLM3 gene encodes a protein that protects against lethal effects of oxidative damage. Cell. Mol. Biol., 47, 1149–1162. - PubMed
- Frentzel S., Pesold-Hurt B., Seelig A. & Kloetzel P.M. ( 1994). 20S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13–16S preproteasome complexes. J. Mol. Biol., 236, 975–981. - PubMed
- Gavin A.-C. et al. . ( 2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous