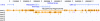Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes - PubMed (original) (raw)
Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes
W James Kent et al. Proc Natl Acad Sci U S A. 2003.
Abstract
This study examines genomic duplications, deletions, and rearrangements that have happened at scales ranging from a single base to complete chromosomes by comparing the mouse and human genomes. From whole-genome sequence alignments, 344 large (>100-kb) blocks of conserved synteny are evident, but these are further fragmented by smaller-scale evolutionary events. Excluding transposon insertions, on average in each megabase of genomic alignment we observe two inversions, 17 duplications (five tandem or nearly tandem), seven transpositions, and 200 deletions of 100 bases or more. This includes 160 inversions and 75 duplications or transpositions of length >100 kb. The frequencies of these smaller events are not substantially higher in finished portions in the assembly. Many of the smaller transpositions are processed pseudogenes; we define a "syntenic" subset of the alignments that excludes these and other small-scale transpositions. These alignments provide evidence that approximately 2% of the genes in the human/mouse common ancestor have been deleted or partially deleted in the mouse. There also appears to be slightly less nontransposon-induced genome duplication in the mouse than in the human lineage. Although some of the events we detect are possibly due to misassemblies or missing data in the current genome sequence or to the limitations of our methods, most are likely to represent genuine evolutionary events. To make these observations, we developed new alignment techniques that can handle large gaps in a robust fashion and discriminate between orthologous and paralogous alignments.
Figures
Fig. 1.
Mouse/human alignments at Actinin α-3 before and after chaining and netting, as displayed at the genome browser at
. The RefSeq genes track shows the exon/intron structure of this human gene, which has an ortholog as well as several paralogs and pseudogenes in the mouse. The all
blastz
Mouse track shows
blastz
alignments colored by mouse chromosome. The orthologous gene is on mouse chromosome 19, which is colored purple. Although
blastz
finds the homology in a very sensitive manner, it is fragmented. The chained
blastz
track shows the alignments after chaining. The chaining links related fragments. The orthologous genes and paralogs are each in a single piece. The chaining also merges some redundant alignments and eliminates a few very low-scoring isolated alignments. The Mouse/Human Alignment Net track is designed to show only the orthologous alignments. In this case, there has been no rearrangement other than moderate-sized insertions and deletions, so the net track is quite simple. Clicking on a chain or net track allows the user to open a new browser on the corresponding region in the other species.
Fig. 2.
(a) Small insertions and deletions. (b) Large insertions and deletions and transposons. Counts of genomewide insertions and deletions plotted vs. their size. The blue line shows a combination of human insertions and mouse deletions, whereas the red line shows mouse insertions and human deletions. The vertical scale is the natural logarithm of the number of insertions/deletions of that size. In b, the counts are grouped in bins of 25. The green line shows the percentage of bases in mouse gaps of that size that are covered by human-specific transposons. The peaks in the insertion/deletion graphs appear to be due to transposons.
Fig. 3.
(a) Log histogram of gap frequencies for gaps <50 bases long. (b) Log histogram of gap frequencies for gaps up to 1,500 bases long. (c) Log histogram of gap frequencies for gaps up to 50,000 bases long. Relative frequency of simultaneous and single gaps are shown in both sequences. The horizontal axis is used for gaps in the mouse sequence, which represent either insertions in human or deletions in mouse. The vertical axis is used for gaps in human. The log of the number of simultaneous gaps of a particular size range is converted into a level of gray to create a 2D histogram. The horizontal and vertical axes are not drawn in; the dark lines where the axis would be reflect the log frequencies of gaps that are in only one sequence. (a) Gaps of <50 bases. Gaps that are purely in mouse or purely in human are especially prominent here. (b) Gaps of 10–1,500 bases. The transposon-induced effects in Fig. 1 can also be seen here. Note also the relative concentration near the diagonal for inserts of <200 bases. This occurrence is mostly due to small inversions and locally divergent sequence. (c) Gaps of 1,000–50,000 bases. In this range, the log frequency of simultaneous gaps of a given combined (human and mouse) gap size differs roughly by a constant from the sum of the log frequencies of the individual one-sided gap sizes in each species. In this sense, these longer simultaneous gaps act as if they arise from independent gaps in each individual species.
Fig. 4.
A 15,000-base inversion containing two transcripts and showing chr7:2077222–2497100 in the November 2002 assembly of the human genome. Numerous smaller rearrangements are also visible in the net track in this picture. In some cases, the smaller ones simply represent paralogous mouse regions filling in when the orthologous mouse region is not yet sequenced.
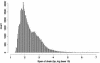
Fig. 5.
Distribution of the spans of all 147,445 chains in the human net. The distribution consists of a bimodal portion for short chains of span <105 and a flat tail for 579 long chains of size between 105 and ≈108.
Comment in
- Chromosome rearrangements in evolution: From gene order to genome sequence and back.
Sankoff D, Nadeau JH. Sankoff D, et al. Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11188-9. doi: 10.1073/pnas.2035002100. Epub 2003 Sep 23. Proc Natl Acad Sci U S A. 2003. PMID: 14506293 Free PMC article. No abstract available.
Similar articles
- Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements.
Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X. Armengol L, et al. Hum Mol Genet. 2003 Sep 1;12(17):2201-8. doi: 10.1093/hmg/ddg223. Epub 2003 Jul 8. Hum Mol Genet. 2003. PMID: 12915466 - Comparative sequence analysis of a single-gene conserved segment in mouse and human.
Thomas JW; NISC Comparative Sequencing Program; Green ED. Thomas JW, et al. Mamm Genome. 2003 Oct;14(10):673-8. doi: 10.1007/s00335-003-2300-1. Mamm Genome. 2003. PMID: 14694903 - Generation of megabase-scale deletions, inversions and duplications involving the Contactin-6 gene in mice by CRISPR/Cas9 technology.
Korablev AN, Serova IA, Serov OL. Korablev AN, et al. BMC Genet. 2017 Dec 28;18(Suppl 1):112. doi: 10.1186/s12863-017-0582-7. BMC Genet. 2017. PMID: 29297312 Free PMC article. - Segmental duplication density decrease with distance to human-mouse breaks of synteny.
Sainz J, Rovensky P, Gudjonsson SA, Thorleifsson G, Stefansson K, Gulcher JR. Sainz J, et al. Eur J Hum Genet. 2006 Feb;14(2):216-21. doi: 10.1038/sj.ejhg.5201534. Eur J Hum Genet. 2006. PMID: 16306878 - Analysis of gene order evolution beyond single-copy genes.
El-Mabrouk N, Sankoff D. El-Mabrouk N, et al. Methods Mol Biol. 2012;855:397-429. doi: 10.1007/978-1-61779-582-4_15. Methods Mol Biol. 2012. PMID: 22407718 Review.
Cited by
- Genome-Wide Identification of the Peanut ASR Gene Family and Its Expression Analysis under Abiotic Stress.
Li J, Ma M, Zeng T, Gu L, Zhu B, Wang H, Du X, Zhu X. Li J, et al. Int J Mol Sci. 2024 Oct 13;25(20):11008. doi: 10.3390/ijms252011008. Int J Mol Sci. 2024. PMID: 39456791 Free PMC article. - KegAlign: Optimizing pairwise alignments with diagonal partitioning.
Gulhan AB, Burhans R, Harris R, Kandemir M, Haeussler M, Nekrutenko A. Gulhan AB, et al. bioRxiv [Preprint]. 2024 Sep 3:2024.09.02.610839. doi: 10.1101/2024.09.02.610839. bioRxiv. 2024. PMID: 39282333 Free PMC article. Preprint. - Genome-Wide Identification of NAC Family Genes and Their Expression Analyses in Response to Osmotic Stress in Cannabis sativa L.
Li Q, Zhang H, Yang Y, Tang K, Yang Y, Ouyang W, Du G. Li Q, et al. Int J Mol Sci. 2024 Aug 30;25(17):9466. doi: 10.3390/ijms25179466. Int J Mol Sci. 2024. PMID: 39273412 Free PMC article. - Genomes of diverse Actinidia species provide insights into cis-regulatory motifs and genes associated with critical traits.
Li X, Huo L, Li X, Zhang C, Gu M, Fan J, Xu C, Gong J, Hu X, Zheng Y, Sun X. Li X, et al. BMC Biol. 2024 Sep 11;22(1):200. doi: 10.1186/s12915-024-02002-z. BMC Biol. 2024. PMID: 39256695 Free PMC article. - Genome-wide identification and expression analysis of the SPL gene family and its response to abiotic stress in barley (Hordeum vulgare L.).
He A, Zhou H, Ma C, Bai Q, Yang H, Yao X, Wu W, Xue G, Ruan J. He A, et al. BMC Genomics. 2024 Sep 9;25(1):846. doi: 10.1186/s12864-024-10773-6. BMC Genomics. 2024. PMID: 39251952 Free PMC article.
References
- Haldane, J. B. S. (1932) The Causes of Evolution (Longmans and Green, London).
- Graur, D. & Li, W. H. (2000) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA).
- The Mouse Sequencing Consortium (2002) Nature 420, 520–562. - PubMed
- Tesler, G. (2002) Bioinformatics 18, 492–493. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources