Extensive gene gain associated with adaptive evolution of poxviruses - PubMed (original) (raw)
Extensive gene gain associated with adaptive evolution of poxviruses
Aoife McLysaght et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Previous studies of genome evolution usually have involved one or two genomes and have thus been limited in their ability to detect the direction and rate of evolutionary change. Here, we use complete genome data from 20 poxvirus genomes to build a robust phylogeny of the Poxviridae and to study patterns of genome evolution. We show that, although there has been little gene order evolution, there are substantial differences between poxviruses in terms of genome content. Furthermore, we show that the rate of gene acquisition is not constant over time and that it has increased in the orthopox lineage (which includes smallpox and vaccinia). We also tested for positive selection on 204 groups of genes and show that a disproportionately high proportion of genes in the orthopox clade are under positive selection. The association of an increased rate of gene gain and positive selection is indicative of adaptive genome evolution. Many of the genes involved in these processes are likely to be associated with host-parasite coevolution.
Figures
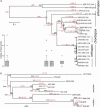
Fig. 1.
NJ trees of the poxviruses. The classifications of the viruses are indicated to the right. Bootstrap values are shown for each branch. The numbers in red separated by/are the number of gain and loss events, respectively that occurred along each branch. At the base of the phylogeny it is not possible to distinguish a gaininone lineage from a loss in the other. In these cases the events were counted as gains and we use a lone hyphen (-) in place of a count of loss events to indicate this fact. *** indicates that the number of gains was significant at the 1% level, and ** indicates significance at the 5% level (see Methods). Gray and black asterisks indicate the number of events were below or above expectations, respectively. (a) NJ tree of all completely sequences poxviruses, based on 34 gene families. The number of genes from each genome that were assigned into families is shown after the genome name. The box plots show the distribution of the number of loss events in families dating back to the node of the tree above (indicated by a labeled circle). Only box plots with outliers are shown. The leftmost box plot refers to families ancestral to all poxvirus genomes analyzed. (b) NJ tree of the orthopoxviruses, based on 92 gene families. The dashed line shows the approximate position of the root of the orthopoxviruses inferred from tree a.
Fig. 2.
The genomes of all 20 poxviruses showing the taxonomic breadth and the genome arrangement of 92 gene families used in the phylogeny inference. Genomes are shown in the same order as in Fig. 1_a_, and names are shown on the right. All genes are indicated by black outline boxes. Horizontal distances are proportional to base pair distances. The 34 orthologs present in all poxviruses are shown in red. The 29 orthologs present in all of the chordopox genomes, but not in the entomopox genomes, are shown in blue. The 29 orthologs present in all orthopox viruses are shown in yellow. Lines along the vertical dimension link orthologs. The orthologs represented are only those that were present in all of the genomes in each classification [i.e., in all pox genomes (red); in all chordopox genomes (blue); or in all orthopox genomes (yellow)].
Similar articles
- Poxvirus genome evolution by gene gain and loss.
Hughes AL, Friedman R. Hughes AL, et al. Mol Phylogenet Evol. 2005 Apr;35(1):186-95. doi: 10.1016/j.ympev.2004.12.008. Epub 2005 Jan 21. Mol Phylogenet Evol. 2005. PMID: 15737590 - A survey of host range genes in poxvirus genomes.
Bratke KA, McLysaght A, Rothenburg S. Bratke KA, et al. Infect Genet Evol. 2013 Mar;14:406-25. doi: 10.1016/j.meegid.2012.12.002. Epub 2012 Dec 23. Infect Genet Evol. 2013. PMID: 23268114 Free PMC article. - Poxviruses: past, present and future.
Lefkowitz EJ, Wang C, Upton C. Lefkowitz EJ, et al. Virus Res. 2006 Apr;117(1):105-18. doi: 10.1016/j.virusres.2006.01.016. Epub 2006 Feb 24. Virus Res. 2006. PMID: 16503070 Review. - Poxvirus Host Range Genes and Virus-Host Spectrum: A Critical Review.
Oliveira GP, Rodrigues RAL, Lima MT, Drumond BP, Abrahão JS. Oliveira GP, et al. Viruses. 2017 Nov 7;9(11):331. doi: 10.3390/v9110331. Viruses. 2017. PMID: 29112165 Free PMC article. Review. - Prediction of steps in the evolution of variola virus host range.
Smithson C, Purdy A, Verster AJ, Upton C. Smithson C, et al. PLoS One. 2014 Mar 13;9(3):e91520. doi: 10.1371/journal.pone.0091520. eCollection 2014. PLoS One. 2014. PMID: 24626337 Free PMC article.
Cited by
- Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution.
Hahn MW. Hahn MW. Genome Biol. 2007;8(7):R141. doi: 10.1186/gb-2007-8-7-r141. Genome Biol. 2007. PMID: 17634151 Free PMC article. - Whole Genome Characterization of Orthopoxvirus (OPV) Abatino, a Zoonotic Virus Representing a Putative Novel Clade of Old World Orthopoxviruses.
Gruber CEM, Giombini E, Selleri M, Tausch SH, Andrusch A, Tyshaieva A, Cardeti G, Lorenzetti R, De Marco L, Carletti F, Nitsche A, Capobianchi MR, Ippolito G, Autorino GL, Castilletti C. Gruber CEM, et al. Viruses. 2018 Oct 6;10(10):546. doi: 10.3390/v10100546. Viruses. 2018. PMID: 30301229 Free PMC article. - Complete genome sequence of Courdo11 virus, a member of the family Mimiviridae.
Yoosuf N, Pagnier I, Fournous G, Robert C, La Scola B, Raoult D, Colson P. Yoosuf N, et al. Virus Genes. 2014 Apr;48(2):218-23. doi: 10.1007/s11262-013-1016-x. Epub 2013 Dec 1. Virus Genes. 2014. PMID: 24293219 - Extensive ITR expansion of the 2022 Mpox virus genome through gene duplication and gene loss.
Brinkmann A, Kohl C, Pape K, Bourquain D, Thürmer A, Michel J, Schaade L, Nitsche A. Brinkmann A, et al. Virus Genes. 2023 Aug;59(4):532-540. doi: 10.1007/s11262-023-02002-1. Epub 2023 May 31. Virus Genes. 2023. PMID: 37256469 Free PMC article. - Orthopoxvirus Genome Sequencing, Assembly, and Analysis.
Gigante CM, Weigand MR, Li Y. Gigante CM, et al. Methods Mol Biol. 2025;2860:39-63. doi: 10.1007/978-1-0716-4160-6_4. Methods Mol Biol. 2025. PMID: 39621260
References
- Wolfe, K. H. & Li, W. H. (2003) Nat. Genet. 33, 255–265. - PubMed
- McLean, M. J., Wolfe, K. H. & Devine, K. M. (1998) J. Mol. Evol. 47, 691–696. - PubMed
- Sharp, P. M. & Li, W. H. (1986) J. Mol. Evol. 24, 28–38. - PubMed
- Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708–713. - PubMed
- Vision, T. J., Brown, D. G. & Tanksley, S. D. (2000) Science 290, 2114–2117. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources