Identification and characterization of the binding sites of P-glycoprotein for multidrug resistance-related drugs and modulators - PubMed (original) (raw)
Review
Identification and characterization of the binding sites of P-glycoprotein for multidrug resistance-related drugs and modulators
Ahmad R Safa. Curr Med Chem Anticancer Agents. 2004 Jan.
Abstract
A major problem in cancer treatment is the development of resistance to multiple chemotherapeutic agents in tumor cells. A major mechanism of this multidrug resistance (MDR) is overexpression of the MDR1 product P-glycoprotein, known to bind to and transport a wide variety of agents. This review concentrates on the progress made toward understanding the role of this protein in MDR, identifying and characterizing the drug binding sites of P-glycoprotein, and modulating MDR by P-glycoprotein-specific inhibitors. Since our initial discovery that P-glycoprotein binds to vinblastine photoaffinity analogs, many P-glycoprotein-specific photoaffinity analogs have been developed and used to identify the particular domains of P-glycoprotein capable of interacting with these analogs and other P-glycoprotein substrates. Furthermore, significant advances have been made in delineating the drug binding sites of this protein by studying mutant P-glycoprotein. Photoaffinity labeling experiments and the use of site-directed antibodies to several domains of this protein have allowed the localization of the general binding domains of some of the cytotoxic agents and MDR modulators on P-glycoprotein. Moreover, site-directed mutagenesis studies have identified the amino acids critical for the binding of some of these agents to P-glycoprotein. Furthermore, equilibrium binding assays using plasma membranes from MDR cells and radioactive drugs have aided our understanding of the modes of drug interactions with P-glycoprotein. Based on the available data, a topological model of P-glycoprotein and the approximate locations of its drug binding sites, as well as a proposed classification of multiple drug binding sites of this protein, is presented in this review.
Figures
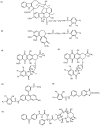
Fig. (1)
Structures of photoaffinity analogs of MDR-related drugs known to covalently label P-glycoprotein: (I) [125I]NASV, (2) [125I]NASC, (3) [3H]NAB-DNR, (4) [125I]NAS-DNR, (5) [125I]ASA-Rh123, (6) [125I]ASA-BZ, and (7) [3H]BzDC-paclitaxel.
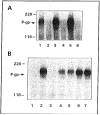
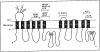
Fig. (2)
(A) Autoradiogram of [125I]NASV photolabeled plasma membranes (20 μg protein) from CEM, CEM/VBL100, CEM/VBL1000 and CEM/VBL5000 cells in the absence (lanes 1, 2, 4 and 6) or presence of 10 μM nonradioactive vinblastine (lanes 3, 5 and 7). (B) Autoradiogram of [125I]NASV photoaffinity labeled KB-3-1 (lane 1) and KB-GRC1 transfectants (KB-3-1 cells transfected with the _MDR_1 gene) (lanes 2–7), KB-GRC1 transfectants (8 × 106 cells) were photoaffinity labeled with 50 nM [125I]NASV (50–60 Ci/mMole) in the absence (lane 2) or presence of 100 μM vinblastine, actinomycin D, doxorubicin, colchicine, or methotrexate, respectively (lanes 3–7), and after subjecting to SDS-PAGE, the samples were processed for autoradiography.
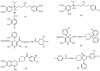
Fig. (3)
Structures of photoaffinity analogs of MDR modulators known to covalently label P-glycoprotein: (1) [125I]NAS-VP, (2) LU-49888, (3)[3H]azidopme, (4) [3H]B92009-005, (5) [125I]AAP and (6) [125I]NAPS.
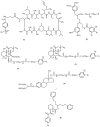
Fig. (4)
Structures of photoaffinity analogs of MDR modulators known to covalently label P-glycoprotein: (1) cyclosporin aziridine, (2) synthetic isoprenoid, (3) _N_-solanesyl-N', N'-bis(3, 4-dimethoxybenzoyl)ethylenediamine, (4) forskolin analogs, (5) estramustine photoaffinity analog, and (6) VF-13.
Fig. (5)
Topological model of P-glycoprotein and the approximate locations of its drug binding sites. The figure shows the structure of P-glycoprotein with twelve transmembrane domains (TMDs) predicted by hydrophobicity plot. Domains of the protein that are involved in the binding of iodomycin (IDM), [125I]NASV, [125I]NAS-VP, [125I]NAST, [3H]azidopine (AZP), 6-o-[[2-[3-(4-azido-3-[125I]iodophenyl) propionamido]ethyl]-carbamyl)forskolin (AIPPF), [125I]iodoarylazidoprazosin (IAAP), [3H]-3'-BzDC-paclitaxel, [3H]-7-BzDC-paclitaxel (dashed lines, amino acid residues 985–1008 are marked). For details, see the text.
Fig. (6)
Photoaffinity labeling of P-glycoprotein from SH-SY5Y/VCR cells with [125I]AAP (lane 1) and the effects of 100 μM trifluoperazine, chlorpromazine, thioridazine, perphenazine, W-7, cis-flupentixol, trans-flupentixol, fluphenazine and fluphenazine N-mustard (lanes 2–10), respectively.
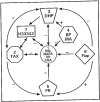
Fig. (7)
Proposed classification of multiple drug binding sites of P-glycoprotein. Based on the kinetic analysis of drug binding to P-glycoprotein under equilibrium conditions and photoaffinity labeling using various photoaffinity analogs of the cytotoxic agents and MDR modulators as well as competition experiments described in the text, seven binding sites for P-glycoprotein are proposed. Arrows indicate positive or negative communication between these sites.
Similar articles
- Photoaffinity analogs for multidrug resistance-related transporters and their use in identifying chemosensitizers.
Safa AR. Safa AR. Drug Resist Updat. 1999 Dec;2(6):371-381. doi: 10.1054/drup.1999.0105. Drug Resist Updat. 1999. PMID: 11498353 - Indoloquinoxaline compounds that selectively antagonize P-glycoprotein.
Smith CD, Myers CB, Zilfou JT, Smith SN, Lawrence DS. Smith CD, et al. Oncol Res. 2000;12(5):219-29. doi: 10.3727/096504001108747710. Oncol Res. 2000. PMID: 11417747 - Celastraceae sesquiterpenes as a new class of modulators that bind specifically to human P-glycoprotein and reverse cellular multidrug resistance.
Muñoz-Martínez F, Lu P, Cortés-Selva F, Pérez-Victoria JM, Jiménez IA, Ravelo AG, Sharom FJ, Gamarro F, Castanys S. Muñoz-Martínez F, et al. Cancer Res. 2004 Oct 1;64(19):7130-8. doi: 10.1158/0008-5472.CAN-04-1005. Cancer Res. 2004. PMID: 15466210 - Modulation and prevention of multidrug resistance by inhibitors of P-glycoprotein.
Sikic BI, Fisher GA, Lum BL, Halsey J, Beketic-Oreskovic L, Chen G. Sikic BI, et al. Cancer Chemother Pharmacol. 1997;40 Suppl:S13-9. doi: 10.1007/s002800051055. Cancer Chemother Pharmacol. 1997. PMID: 9272128 Review. - Pharmacological strategies for overcoming multidrug resistance.
Nobili S, Landini I, Giglioni B, Mini E. Nobili S, et al. Curr Drug Targets. 2006 Jul;7(7):861-79. doi: 10.2174/138945006777709593. Curr Drug Targets. 2006. PMID: 16842217 Review.
Cited by
- Insight into pleiotropic drug resistance ATP-binding cassette pump drug transport through mutagenesis of Cdr1p transmembrane domains.
Rawal MK, Khan MF, Kapoor K, Goyal N, Sen S, Saxena AK, Lynn AM, Tyndall JD, Monk BC, Cannon RD, Komath SS, Prasad R. Rawal MK, et al. J Biol Chem. 2013 Aug 23;288(34):24480-93. doi: 10.1074/jbc.M113.488353. Epub 2013 Jul 3. J Biol Chem. 2013. PMID: 23824183 Free PMC article. - Microscopic Characterization of Membrane Transporter Function by In Silico Modeling and Simulation.
Vermaas JV, Trebesch N, Mayne CG, Thangapandian S, Shekhar M, Mahinthichaichan P, Baylon JL, Jiang T, Wang Y, Muller MP, Shinn E, Zhao Z, Wen PC, Tajkhorshid E. Vermaas JV, et al. Methods Enzymol. 2016;578:373-428. doi: 10.1016/bs.mie.2016.05.042. Epub 2016 Jul 11. Methods Enzymol. 2016. PMID: 27497175 Free PMC article. Review. - Pharmacokinetics of substrate uptake and distribution in murine brain after nasal instillation.
Graff CL, Zhao R, Pollack GM. Graff CL, et al. Pharm Res. 2005 Feb;22(2):235-44. doi: 10.1007/s11095-004-1191-5. Pharm Res. 2005. PMID: 15783071 - Fexofenadine hydrochloride in the treatment of allergic disease: a review.
Axelrod D, Bielory L. Axelrod D, et al. J Asthma Allergy. 2008 Sep 19;1:19-29. doi: 10.2147/jaa.s3092. J Asthma Allergy. 2008. PMID: 21436982 Free PMC article. - The Identification of New c-FLIP Inhibitors for Restoring Apoptosis in TRAIL-Resistant Cancer Cells.
Yaacoub K, Pedeux R, Lafite P, Jarry U, Aci-Sèche S, Bonnet P, Daniellou R, Guillaudeux T. Yaacoub K, et al. Curr Issues Mol Biol. 2024 Jan 12;46(1):710-728. doi: 10.3390/cimb46010046. Curr Issues Mol Biol. 2024. PMID: 38248348 Free PMC article.
References
- Gottesman MM, Fojo T, Bates SE. Nat. Rev. Cancer. 2002;2:48. - PubMed
- Leonard GD, Polgar O, Bates SE. Curr. Opin. Investig. Drugs. 2002;3:1652. - PubMed
- Cole SP, Deely RG. Bioessays. 1998;20:931. - PubMed
- Kruh GD, Zeng H, Rea PA, Liu G, Chen ZS, Lee K, Belinsky MG. J. Bioenerg. Biomembr. 2001;33:493. - PubMed
- Wijnholds J. Novartis Found Symp. 2002;243:69. discussion 80–2,180–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA090878/CA/NCI NIH HHS/United States
- CA47652/CA/NCI NIH HHS/United States
- CA80734/CA/NCI NIH HHS/United States
- CA56078/CA/NCI NIH HHS/United States
- R01 CA080734/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources