Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements - PubMed (original) (raw)
Comparative Study
Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements
Mark A Rubin et al. Am J Pathol. 2004 Mar.
Abstract
Despite years of discovery and attempts at validation, few molecular biomarkers achieve acceptance in the clinical setting. Tissue-based markers evaluated by immunohistochemistry suffer from a high degree of inter- and intraobserver variability. One recent advance in this field that promises to automate this process is the development of AQUA, a molecular-based method of quantitative assessment of protein expression. This system integrates a set of algorithms that allows for the rapid, automated, continuous, and quantitative analysis of tissue samples, including the separation of tumor from stromal elements and the subcellular localization of signals. This study uses the AQUA system to assess a recently described prostate cancer biomarker, alpha-methylacyl-CoA-racemase (AMACR), and to determine the effectiveness of the quantitative measurement of this marker as a means for making the diagnosis of prostate cancer. Using a prostate cancer progression tissue microarray containing a wide range of prostate tissues, AQUA was directly compared to standard immunohistochemical evaluation for AMACR protein expression using the p504s monoclonal antibody. Both methods produced similar results showing AMACR protein expression to be strongest in the clinically localized prostate cancer, followed by the metastatic tumor samples. Benign prostate tissue was categorized as negative for most tissue samples by immunohistochemistry. However, AMACR was detectable using the AQUA system at low levels using the standard 1:25 dilution but also at 1:250 dilution, which is not detectable by light microscopy. The AQUA system was also able to discriminate foamy gland prostate cancers, which are known to have a lower AMACR expression than typical acinar prostate cancers, from benign prostate tissue samples. Finally, a receiver-operating-characteristic curve was plotted to determine the specificity of the AMACR AQUA Z-score (normalized AQUA score) to predict that a given tissue microarray sample contains cancer. The area under the curve was calculated at 0.90 (P < 0.00001; 95% CI, 0.84 to 0.95). At an AMACR AQUA Z-score score of -0.3, 91% of the 70 samples classified as prostate cancer were correctly categorized without the intervention of a pathologist reviewing the tissue microarray slide. In conclusion, the AQUA system provides a continuous measurement of AMACR on a wide range of prostate tissue samples. In the future, the AMACR AQUA Z-score may be useful in the automated screening and evaluation of prostate tissue biomarkers.
Figures
Figure 1
A–D: Prostate cancer progression chip to measure AMACR expression in benign and neoplastic prostate tissue samples. A prostate cancer progression tissue microarray was designed composed of benign prostate sectors (dark blue), clinically localized prostate cancer sectors (red), regional lymph node metastatic prostate tumors (green), and distant hormone-refractory metastatic prostate tumors (light blue). A: AMACR expression can be seen at the macroscopic level with strong brown expression seen in the prostate cancer sectors. Rare focal expression can be appreciated in the benign section (blue square and TMA sample) and at a similar intensity as seen in an example of clinically localized prostate cancer (red square and TMA sample). C: This sample demonstrates high-grade PIN, a precursor lesion to prostate cancer. D: This area of high-grade PIN demonstrates moderate levels of AMACR expression. Original magnifications: ×200 (A, B); ×630 (C, D).
Figure 2
A–H: Schematic view for using AQUA to measure AMACR in prostate samples. A: Prostate cancers are heterogeneous with a mixture of benign and infiltrating cancer cells. B: Using a single antibody against keratin, the AQUA algorithm can measure the intensity and area of keratin-positive cells using fluorescent dyes. This area is shown in yellow. C: A second antibody against AMACR (p504s) is used to target neoplastic prostate glands. This area is shown in red. D: The fluorescent dyes allow for the simultaneous evaluation of up to four antibodies. In this example AMACR and keratin are shown. The area for each marker is measured by the system (E and F) and can be considered a virtual compartment representing a keratin-positive area and AMACR-positive area (G). Dual expression of keratin and AMACR define a neoplastic compartment (ie, prostate cancer or prostatic intraepithelial neoplasia). Keratin-positive and AMACR-negative areas are consistent with benign epithelial compartment. Negative staining for both AMACR and keratin are consistent with a stromal compartment. Using these definitions, the intensities of different compartments can be measured using arbitrary units (H).
Figure 3
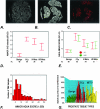
A–G: AMACR expression as determined by AQUA and standard pathology review. A to C demonstrate how AQUA identifies prostate cancer from benign tissue in a tissue microarray sample (0.6 mm diameter). A: Using DAPI staining, all nucleated cells in both the stroma and epithelial compartments are identified. The cytokeratin-positive compartment identifies all epithelial cells (B) and Cy-5 expression (red) demonstrates the area of AMACR expression (C). The staining area and intensity of each compartment can be calculated. The AMACR-positive area, which overlaps with the cytokeratin mask is equivalent to the prostate cancer compartment. D: AMACR (p504s) protein expression by standard pathology review using immunohistochemistry. The expression of AMACR (p504s) was determined using the prostate cancer progression tissue microarray. Expression was scored as negative (score = 1), weak (score = 2), moderate (score = 3), and strong (score = 4). The results for each tissue type are presented as error bars of AMACR (p504s) protein expression with 95% confidence intervals. E: AQUA evaluation of AMACR expression using the prostate cancer progression tissue microarray at two antibody concentrations. AQUA measured similar AMACR protein expression levels for the different tissue categories [ie, benign, localized prostate cancer (PCa), metastatic prostate cancer to regional lymph nodes (Mets), and distant hormone-refractory metastatic prostate cancer (HR Mets)] at standard antibody concentrations (1:25) and at a concentration that would not be visible using standard immunohistochemistry (1:250). The intensity is corrected for area for each tissue microarray spot and measured using arbitrary units. The errors bars demonstrate that within 95% confidence intervals, benign prostate tissue does not score greater than 10 units. F: AMACR protein expression histogram for AQUA analysis. Using the arbitrary unit, this histogram demonstrates a relatively normal distribution of AMACR protein expression using samples from the prostate progression chip. This demonstrates the ability of the AQUA system to give a continuous readout of protein expression. G: Variable AMACR protein expression is appreciated by presenting the individual staining intensities for each sample based on tissue category. Although AMACR protein expression for the clinically localized prostate tumors is significantly higher than the population of benign samples, AMACR expression is still observed in benign tissues and can also be found to be low in other categories.
Figure 4
Evaluation of normalized AMACR-AQUA scores and ability to predict the presence of prostate cancer. The AQUA results were normalized creating an AMACR AQUA _Z_-score (left). Expression results demonstrate measurable levels of AMACR in the foamy gland tumor population that are statistically significantly higher than the benign prostate tissue population as demonstrated by nonoverlapping error bars with 95% confidence intervals. A ROC plot demonstrates the sensitivity and specificity of using an AMACR AQUA _Z_-score to diagnose prostate cancer in an unsupervised manner (ie, without previous review by a pathologist) (right). The area under the ROC curve is 0.90 (P < 0.00001; 95% CI, 0.84 to 0.95). An area under the curve of 1.00 represents perfect discrimination of the test to predict prostate cancer and 0.50 no discriminatory power.
Similar articles
- Expression and diagnostic utility of alpha-methylacyl-CoA-racemase (P504S) in foamy gland and pseudohyperplastic prostate cancer.
Zhou M, Jiang Z, Epstein JI. Zhou M, et al. Am J Surg Pathol. 2003 Jun;27(6):772-8. doi: 10.1097/00000478-200306000-00007. Am J Surg Pathol. 2003. PMID: 12766580 - alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer.
Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. Rubin MA, et al. JAMA. 2002 Apr 3;287(13):1662-70. doi: 10.1001/jama.287.13.1662. JAMA. 2002. PMID: 11926890 - Foamy gland carcinoma of the prostate in needle biopsy: incidence, Gleason grade, and comparative α-methylacyl-CoA racemase vs. ERG expression.
Warrick JI, Humphrey PA. Warrick JI, et al. Am J Surg Pathol. 2013 Nov;37(11):1709-14. doi: 10.1097/PAS.0b013e318293d85b. Am J Surg Pathol. 2013. PMID: 23797726 - Discovery and clinical application of a novel prostate cancer marker: alpha-methylacyl CoA racemase (P504S).
Jiang Z, Woda BA, Wu CL, Yang XJ. Jiang Z, et al. Am J Clin Pathol. 2004 Aug;122(2):275-89. doi: 10.1309/EJUY-UQPE-X1MG-68MK. Am J Clin Pathol. 2004. PMID: 15323145 Review. - Application of alpha-methylacyl coenzyme A racemase immunohistochemistry in the diagnosis of prostate cancer: a review.
Adley BP, Yang XJ. Adley BP, et al. Anal Quant Cytol Histol. 2006 Feb;28(1):1-13. Anal Quant Cytol Histol. 2006. PMID: 16566275 Review.
Cited by
- Quantitative analysis of BTF3, HINT1, NDRG1 and ODC1 protein over-expression in human prostate cancer tissue.
Symes AJ, Eilertsen M, Millar M, Nariculam J, Freeman A, Notara M, Feneley MR, Patel HR, Masters JR, Ahmed A. Symes AJ, et al. PLoS One. 2013 Dec 27;8(12):e84295. doi: 10.1371/journal.pone.0084295. eCollection 2013. PLoS One. 2013. PMID: 24386364 Free PMC article. - Cancer immunology--analysis of host and tumor factors for personalized medicine.
Ogino S, Galon J, Fuchs CS, Dranoff G. Ogino S, et al. Nat Rev Clin Oncol. 2011 Aug 9;8(12):711-9. doi: 10.1038/nrclinonc.2011.122. Nat Rev Clin Oncol. 2011. PMID: 21826083 Free PMC article. Review. - Quantification of diverse subcellular immunohistochemical markers with clinicobiological relevancies: validation of a new computer-assisted image analysis procedure.
Lejeune M, Jaén J, Pons L, López C, Salvadó MT, Bosch R, García M, Escrivà P, Baucells J, Cugat X, Alvaro T. Lejeune M, et al. J Anat. 2008 Jun;212(6):868-78. doi: 10.1111/j.1469-7580.2008.00910.x. J Anat. 2008. PMID: 18510512 Free PMC article. - Histo-ELISA technique for quantification and localization of tissue components.
Li Z, Goebel S, Reimann A, Ungerer M. Li Z, et al. Sci Rep. 2020 Nov 16;10(1):19849. doi: 10.1038/s41598-020-76950-1. Sci Rep. 2020. PMID: 33199754 Free PMC article. - Defining aggressive prostate cancer using a 12-gene model.
Bismar TA, Demichelis F, Riva A, Kim R, Varambally S, He L, Kutok J, Aster JC, Tang J, Kuefer R, Hofer MD, Febbo PG, Chinnaiyan AM, Rubin MA. Bismar TA, et al. Neoplasia. 2006 Jan;8(1):59-68. doi: 10.1593/neo.05664. Neoplasia. 2006. PMID: 16533427 Free PMC article.
References
- Dowsett M, Bartlett J, Ellis IO, Salter J, Hills M, Mallon E, Watters AD, Cooke T, Paish C, Wencyk PM, Pinder SE. Correlation between immunohistochemistry (HercepTest) and fluorescence in situ hybridization (FISH) for HER-2 in 426 breast carcinomas from 37 centres. J Pathol. 2003;199:418–423. - PubMed
- Paik S, Bryant J, Tan-Chiu E, Romond E, Hiller W, Park K, Brown A, Yothers G, Anderson S, Smith R, Wickerham DL, Wolmark N. Real-world performance of HER2 testing—National Surgical Adjuvant Breast and Bowel Project experience. J Natl Cancer Inst. 2002;94:852–854. - PubMed
- Fernandez PL, Arce Y, Farre X, Martinez A, Nadal A, Rey MJ, Peiro N, Campo E, Cardesa A. Expression of p27/Kip1 is down-regulated in human prostate carcinoma progression. J Pathol. 1999;187:563–566. - PubMed
- Yang RM, Naitoh J, Murphy M, Wang HJ, Phillipson J, deKernion JB, Loda M, Reiter RE. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159:941–945. - PubMed
- Guo Y, Sklar GN, Borkowski A, Kyprianou N. Loss of the cyclin-dependent kinase inhibitor p27 (Kip1) protein in human prostate cancer correlates with tumor grade. Clin Cancer Res. 1997;3:2269–2274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AG21404/AG/NIA NIH HHS/United States
- P50 CA090381/CA/NCI NIH HHS/United States
- R01 AG021404/AG/NIA NIH HHS/United States
- P50CA69568/CA/NCI NIH HHS/United States
- P50CA90381/CA/NCI NIH HHS/United States
- R21 CA100825/CA/NCI NIH HHS/United States
- P50 CA069568/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical