Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia - PubMed (original) (raw)
doi: 10.1128/JVI.79.2.884-895.2005.
Susanna K P Lau, Chung-ming Chu, Kwok-hung Chan, Hoi-wah Tsoi, Yi Huang, Beatrice H L Wong, Rosana W S Poon, James J Cai, Wei-kwang Luk, Leo L M Poon, Samson S Y Wong, Yi Guan, J S Malik Peiris, Kwok-yung Yuen
Affiliations
- PMID: 15613317
- PMCID: PMC538593
- DOI: 10.1128/JVI.79.2.884-895.2005
Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia
Patrick C Y Woo et al. J Virol. 2005 Jan.
Abstract
Despite extensive laboratory investigations in patients with respiratory tract infections, no microbiological cause can be identified in a significant proportion of patients. In the past 3 years, several novel respiratory viruses, including human metapneumovirus, severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV), and human coronavirus NL63, were discovered. Here we report the discovery of another novel coronavirus, coronavirus HKU1 (CoV-HKU1), from a 71-year-old man with pneumonia who had just returned from Shenzhen, China. Quantitative reverse transcription-PCR showed that the amount of CoV-HKU1 RNA was 8.5 to 9.6 x 10(6) copies per ml in his nasopharyngeal aspirates (NPAs) during the first week of the illness and dropped progressively to undetectable levels in subsequent weeks. He developed increasing serum levels of specific antibodies against the recombinant nucleocapsid protein of CoV-HKU1, with immunoglobulin M (IgM) titers of 1:20, 1:40, and 1:80 and IgG titers of <1:1,000, 1:2,000, and 1:8,000 in the first, second and fourth weeks of the illness, respectively. Isolation of the virus by using various cell lines, mixed neuron-glia culture, and intracerebral inoculation of suckling mice was unsuccessful. The complete genome sequence of CoV-HKU1 is a 29,926-nucleotide, polyadenylated RNA, with G+C content of 32%, the lowest among all known coronaviruses with available genome sequence. Phylogenetic analysis reveals that CoV-HKU1 is a new group 2 coronavirus. Screening of 400 NPAs, negative for SARS-CoV, from patients with respiratory illness during the SARS period identified the presence of CoV-HKU1 RNA in an additional specimen, with a viral load of 1.13 x 10(6) copies per ml, from a 35-year-old woman with pneumonia. Our data support the existence of a novel group 2 coronavirus associated with pneumonia in humans.
Figures
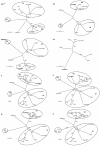
FIG. 1.
Genome organization of CoV-HKU1. Overall organization of the 29,926-nucleotide CoV-HKU1 genomic RNA. Predicted ORFs 1a and 1b, encoding the nonstructural polyproteins (p28, p65, and nsp1 to -13) and those encoding the hemagglutinin-esterase, spike, envelope, membrane and nucleocapsid structural proteins are indicated. Arrows indicate putative cleavage sites (with the corresponding nucleotide positions) of the replicase polyprotein encoded by ORF 1a and ORF 1b. ATR and PL1pro and PL2pro represent the acidic tandem repeat and the two papain-like proteases, respectively, in nsp1.
FIG. 2.
Phylogenetic analysis of chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase (Pol), helicase, hemagglutinin-esterase (HE), spike (S), envelope (E), membrane (M), and nucleocapsid (N) of CoV-HKU1. The trees were constructed by the neighbor-joining method, using Jukes-Cantor correction and bootstrap values calculated from 1,000 trees. Three hundred three, 928, 595, 418, 1356, 75, 225, and 406 amino acid positions in 3CLpro, Pol, helicase, HE, S, E, M and N, respectively, were included in the analysis. The scale bar indicates the estimated number of substitutions per 10 amino acids. HCoV-229E, human coronavirus 229E; PEDV, porcine epidemic diarrhea virus; PTGV, porcine transmissible gastroenteritis virus; CCoV, canine enteric coronavirus; HCoV-NL63, human coronavirus NL63; HCoV-OC43, human coronavirus OC43; MHV, murine hepatitis virus; BCoV, bovine coronavirus; SDAV, rat sialodacryoadenitis coronavirus; ECoV, equine coronavirus NC99; PHEV, porcine hemagglutinating encephalomyelitis virus; IBV, infectious bronchitis virus; SARS-CoV, SARS coronavirus.
FIG. 3.
Arrangements of proteins in replicase polyprotein in HKU1 compared with those in HCoV-OC43, BCoV, and MHV. Alignment of the AC domains of HCoV-OC43, BCoV, and MHV and the AC domains and ATR (underlined) of CoV-HKU1 in the two patients was generated with ClustalX 1.83. AC domain, acidic domain. GenBank accession numbers are as follows: MHV, NC_001846; BCoV, NC_003045; HCoV-OC43, AY585229.
FIG. 4.
Spike protein of CoV-HKU1. The spike protein (1,356 amino acids) of CoV-HKU1 is depicted by the horizontal bar. SS, N terminal signal sequence (amino acid residues 1 to 13); HR1, heptad repeat 1 (amino acid residues 982 to 1083); HR2, heptad repeat 2 (amino acid residues 1250 to 1297); TM, transmembrane domain (amino acid residues 1301 to 1323). Alignment of the N-terminal region important for receptor binding (amino acid residues 1 to 330) and the region upstream of the cleavage site between S1 and S2 of CoV-HKU1 and other group 2 coronaviruses was done with ClustalX 1.83. Residues that match the CoV-HKU1 sequence exactly are boxed. The three conserved regions (sites I, II, and III) for receptor binding in MHV are shaded. The positions of the four conserved amino acids important for receptor binding in MHV are indicated with arrows. GenBank accession numbers were as follows: MHV, P11224; BCoV, NP_150077; HCoV-OC43, NP_937950; SDAV, AAF97738; PHEV, AAL80031; ECoV, AAQ67205.
FIG. 5.
Sequential quantitative RT-PCR for CoV-HKU1 in NPAs and serum IgG titers against N protein of CoV-HKU1.
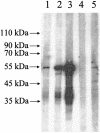
FIG. 6.
Western blot analysis of purified recombinant CoV-HKU1 N protein antigen. Prominent immunoreactive protein bands of about 53 kDa were visible on the Western blot that used recombinant N protein as the antigen during the second and fourth weeks of the patient's illness (lanes 2 and 3). Only very faint bands were observed for serum samples obtained from the patient during the first week of the illness (lane 1) and two healthy blood donors (lanes 4 and 5).
Similar articles
- Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia.
Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, Chu CM, Lee RA, Luk WK, Wong GK, Wong BH, Cheng VC, Tang BS, Wu AK, Yung RW, Chen H, Guan Y, Chan KH, Yuen KY. Woo PC, et al. J Infect Dis. 2005 Dec 1;192(11):1898-907. doi: 10.1086/497151. Epub 2005 Oct 20. J Infect Dis. 2005. PMID: 16267760 Free PMC article. - Coronavirus HKU1 and other coronavirus infections in Hong Kong.
Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, Lee P, Tang BS, Cheung CH, Lee RA, So LY, Lau YL, Chan KH, Yuen KY. Lau SK, et al. J Clin Microbiol. 2006 Jun;44(6):2063-71. doi: 10.1128/JCM.02614-05. J Clin Microbiol. 2006. PMID: 16757599 Free PMC article. - Two cases of severe obstructive pneumonia associated with an HKU1-like coronavirus.
Kupfer B, Simon A, Jonassen CM, Viazov S, Ditt V, Tillmann RL, Müller A, Matz B, Schildgen O. Kupfer B, et al. Eur J Med Res. 2007 Mar 26;12(3):134-8. Eur J Med Res. 2007. PMID: 17507310 - Coronavirus diversity, phylogeny and interspecies jumping.
Woo PC, Lau SK, Huang Y, Yuen KY. Woo PC, et al. Exp Biol Med (Maywood). 2009 Oct;234(10):1117-27. doi: 10.3181/0903-MR-94. Epub 2009 Jun 22. Exp Biol Med (Maywood). 2009. PMID: 19546349 Review. - The widening scope of coronaviruses.
Kahn JS. Kahn JS. Curr Opin Pediatr. 2006 Feb;18(1):42-7. doi: 10.1097/01.mop.0000192520.48411.fa. Curr Opin Pediatr. 2006. PMID: 16470161 Review.
Cited by
- Insights on the Structural Variations of the Furin-Like Cleavage Site Found Among the December 2019-July 2020 SARS-CoV-2 Spike Glycoprotein: A Computational Study Linking Viral Evolution and Infection.
Cueno ME, Ueno M, Iguchi R, Harada T, Miki Y, Yasumaru K, Kiso N, Wada K, Baba K, Imai K. Cueno ME, et al. Front Med (Lausanne). 2021 Mar 10;8:613412. doi: 10.3389/fmed.2021.613412. eCollection 2021. Front Med (Lausanne). 2021. PMID: 33777970 Free PMC article. - Multicenter case-control study protocol of pneumonia etiology in children: Global Approach to Biological Research, Infectious diseases and Epidemics in Low-income countries (GABRIEL network).
Picot VS, Bénet T, Messaoudi M, Telles JN, Chou M, Eap T, Wang J, Shen K, Pape JW, Rouzier V, Awasthi S, Pandey N, Bavdekar A, Sanghvi S, Robinson A, Contamin B, Hoffmann J, Sylla M, Diallo S, Nymadawa P, Dash-Yandag B, Russomando G, Basualdo W, Siqueira MM, Barreto P, Komurian-Pradel F, Vernet G, Endtz H, Vanhems P, Paranhos-Baccalà G; pneumonia GABRIEL network. Picot VS, et al. BMC Infect Dis. 2014 Dec 10;14:635. doi: 10.1186/s12879-014-0635-8. BMC Infect Dis. 2014. PMID: 25927410 Free PMC article. - Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A.
Lau SK, Woo PC, Li KS, Tsang AK, Fan RY, Luk HK, Cai JP, Chan KH, Zheng BJ, Wang M, Yuen KY. Lau SK, et al. J Virol. 2015 Mar;89(6):3076-92. doi: 10.1128/JVI.02420-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552712 Free PMC article. - Broad strategies for neutralizing SARS-CoV-2 and other human coronaviruses with monoclonal antibodies.
Ling Z, Yi C, Sun X, Yang Z, Sun B. Ling Z, et al. Sci China Life Sci. 2023 Apr;66(4):658-678. doi: 10.1007/s11427-022-2215-6. Epub 2022 Nov 24. Sci China Life Sci. 2023. PMID: 36443513 Free PMC article. Review. - HexaPrime: a novel method for detection of coronaviruses.
Pyrc K, Stożek K, Galan W, Potempa J. Pyrc K, et al. J Virol Methods. 2013 Mar;188(1-2):29-36. doi: 10.1016/j.jviromet.2012.11.039. Epub 2012 Dec 7. J Virol Methods. 2013. PMID: 23219933 Free PMC article.
References
- Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. Sigrist, and E. M. Zdobnov. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. - PMC - PubMed
- Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. - PubMed
- Eickmann, M., S. Becker, H. D. Klenk, H. W. Doerr, K. Stadler, S. Censini, S. Guidotti, V. Masignani, M. Scarselli, M. Mora, C. Donati, J. H. Han, H. C. Song, S. Abrignani, A. Covacci, and R. Rappuoli. 2003. Phylogeny of the SARS coronavirus. Science 302:1504-1505. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous