MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A - PubMed (original) (raw)
MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A
Amy E Loercher et al. J Cell Biol. 2005.
Abstract
Cell cycle exit is required for proper differentiation in most cells and is critical for normal development, tissue homeostasis, and tumor suppression. However, the mechanisms that link cell cycle exit with differentiation remain poorly understood. Here, we show that the master melanocyte differentiation factor, microphthalmia transcription factor (MITF), regulates cell cycle exit by activating the cell cycle inhibitor INK4A, a tumor suppressor that frequently is mutated in melanomas. MITF binds the INK4A promoter, activates p16(Ink4a) mRNA and protein expression, and induces retinoblastoma protein hypophosphorylation, thereby triggering cell cycle arrest. This activation of INK4A was required for efficient melanocyte differentiation. Interestingly, MITF was also required for maintaining INK4A expression in mature melanocytes, creating a selective pressure to escape growth inhibition by inactivating INK4A. These findings demonstrate that INK4A can be regulated by a differentiation factor, establish a mechanistic link between melanocyte differentiation and cell cycle exit, and potentially explain the tissue-specific tendency for INK4A mutations to occur in melanoma.
Figures
Figure 1.
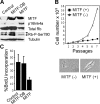
MITF inhibits cell proliferation. (A) Western blot demonstrating MITF, p16Ink4a, total Rb, Rb-phospho-Ser780, and tubulin protein expression in 10T1/2 mouse fibroblasts stably expressing ectopic MITF, MITF-DB (DNA-binding mutant), or empty control vector. Arrows indicate hyperphosphorylated (top band) and hypophosphorylated (bottom band) forms of Rb. (B) Growth curves of 10T1/2 cells stably expressing MITF or empty control vector. Panels show morphology of 10T1/2 cells expressing control empty vector (left) and MITF (right). (C) BrdU incorporation assays in 10T1/2 cells expressing ectopic MITF , MITF-DB, or empty control vector.
Figure 2.
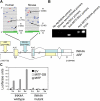
MITF activates the INK4A gene. (A) The ARF/INK4A locus (also called CDKN2A) and location of MITF-binding sites in the INK4A promoter. Exons 1α, 1β, 2, and 3 are indicated, along with their contribution to the INK4A and ARF mRNA transcripts. The nucleotide positions, relative to the centromere, are indicated. The INK4A promoter region is located upstream of exon 1α and downstream of exon 1β, which is transcribed only in the p14ARF transcript. A distinct ARF promoter is located upstream of exon 1β. Above the locus map, the INK4A promoter is depicted in greater detail for both human and mouse. Each line represents 70 bases of FASTA sequence from the NCBI web site (
). Blue bars represent E boxes and red bars M boxes. The E box and M box nearest to the start site in exon 1α (green lines) are indicated. The direction of transcription is indicated by green arrows. (B) Chromatin immunoprecipitation assay in normal melanocytes. Chromatin was immunoprecipitated with an anti-MITF antibody or control mouse IgG, and then was PCR amplified using primers for the INK4A promoter or the SILV gene promoter, a known MITF target. (C) Luciferase assays in 10T1/2 cells transfected with expression vectors for MITF, MITF-DB, or an empty vector control (EV), along with a luciferase reporter driven by the INK4A promoter or a mutant INK4A promoter containing two nucleotide substitutions in the M box (1x, 2x, and 3x = 1-, 2-, and 3-μg plasmid DNA, respectively).
Figure 3.
MITF regulates p16 Ink4a and Rb in normal melanocytes, but not in melanoma cells. (A) Real-time PCR assays of MITF and INK4A mRNA levels in 10T1/2 cells expressing ectopic MITF or an empty control vector (left panels) and in UM41 normal melanocytes transfected with MITF siRNA or control scrambled siRNA (right panels). Expression values represent fold difference compared with GAPDH. (B) Western blot analysis of MITF, p16Ink4a, total Rb, Rb-phospho-Ser780, and tubulin protein expression in UM41 cells 48 h after transfection with plasmids expressing MITF siRNA or control scrambled siRNA. Arrows indicate hyperphosphorylated (top band) and hypophosphorylated (bottom band) forms of Rb. (C) BrdU incorporation assays in UM41 cells transfected with MITF siRNA or control scrambled siRNA. (D) Real-time PCR assays of MITF and p16Ink4a mRNA levels in untransfected Mel202 melanoma cells. (E) Western blot analysis of MITF, p16Ink4a, total Rb, Rb-phospho-Ser780, and tubulin protein expression in Mel202 cells 48 h after transfection with plasmids expressing MITF siRNA or control scrambled siRNA. (F) BrdU incorporation assay in Mel202 cells expressing MITF siRNA or control scrambled siRNA.
Figure 4.
Cells that escape MITF-induced growth inhibition exhibit silencing of INK4A. (A) Growth curve of 10T1/2 cells stably expressing ectopic MITF. Growth-inhibited and escape clones are indicated. (B) Real-time PCR measurement of MITF and p16Ink4a mRNA levels in a growth-inhibited clone and an escape clone. Expression values are fold difference compared with GAPDH. (C) Western blot analysis of MITF, p16Ink4a, and tubulin protein expression in a growth-inhibited clone and an escape clone. (D) Methylation-specific PCR of CpG island in the INK4A promoter region in a growth-inhibited clone and an escape clone, as well as in Mel202 cells. “U” indicates unmethylated and “M” indicates methylated primer sets.
Figure 5
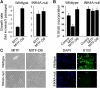
INK4A is required for MITF to execute the melanocyte differentiation program. (A) Growth rate of INK4A-wildtype and INK4A-null MEFs expressing ectopic MITF or MITF-DB. (B) BrdU incorporation assays in INK4A-wildtype and INK4A-null MEFs expressing ectopic MITF, MITF-DB, or empty control vector. (C) Phase-contrast photomicrographs (left panels) of INK4A-wildtype and INK4A-null MEFs expressing ectopic MITF or MITF-DB. Only MITF-expressing wild-type cells exhibit spindle morphology consistent with melanocyte differentiation. Immunofluorescence analysis (right panels) of INK4A-wildtype and INK4A-null MEFs expressing ectopic MITF. DAPI staining (blue) indicates cell nuclei, and green staining indicates expression of the melanocyte marker S100.
Similar articles
- Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression.
Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Carreira S, et al. Nature. 2005 Feb 17;433(7027):764-9. doi: 10.1038/nature03269. Nature. 2005. PMID: 15716956 - Hypoxia-inducible factor 1{alpha} is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells.
Buscà R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, Thyss R, Fitsialos G, Larribère L, Bertolotto C, Virolle T, Barbry P, Pouysségur J, Ponzio G, Ballotti R. Buscà R, et al. J Cell Biol. 2005 Jul 4;170(1):49-59. doi: 10.1083/jcb.200501067. Epub 2005 Jun 27. J Cell Biol. 2005. PMID: 15983061 Free PMC article. - [Malignant melanoma and the role of the paradoxal protein Microphthalmia transcription factor].
Denat L, Larue L. Denat L, et al. Bull Cancer. 2007 Jan;94(1):81-92. Bull Cancer. 2007. PMID: 17237008 Review. French. - Melanocytes and the microphthalmia transcription factor network.
Steingrímsson E, Copeland NG, Jenkins NA. Steingrímsson E, et al. Annu Rev Genet. 2004;38:365-411. doi: 10.1146/annurev.genet.38.072902.092717. Annu Rev Genet. 2004. PMID: 15568981 Review.
Cited by
- Loss of CITED1, an MITF regulator, drives a phenotype switch in vitro and can predict clinical outcome in primary melanoma tumours.
Howlin J, Cirenajwis H, Lettiero B, Staaf J, Lauss M, Saal L, Borg Å, Gruvberger-Saal S, Jönsson G. Howlin J, et al. PeerJ. 2015 Feb 26;3:e788. doi: 10.7717/peerj.788. eCollection 2015. PeerJ. 2015. PMID: 25755924 Free PMC article. - Modulating skin colour: role of the thioredoxin and glutathione systems in regulating melanogenesis.
Lu Y, Tonissen KF, Di Trapani G. Lu Y, et al. Biosci Rep. 2021 May 28;41(5):BSR20210427. doi: 10.1042/BSR20210427. Biosci Rep. 2021. PMID: 33871027 Free PMC article. Review. - [Molecular heterogeneity of malignant melanomas].
Glatz K. Glatz K. Pathologe. 2007 Nov;28(6):474-8. doi: 10.1007/s00292-007-0942-6. Pathologe. 2007. PMID: 17885757 German. - Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation.
Wellbrock C, Marais R. Wellbrock C, et al. J Cell Biol. 2005 Aug 29;170(5):703-8. doi: 10.1083/jcb.200505059. J Cell Biol. 2005. PMID: 16129781 Free PMC article. - Zeb1 represses Mitf and regulates pigment synthesis, cell proliferation, and epithelial morphology.
Liu Y, Ye F, Li Q, Tamiya S, Darling DS, Kaplan HJ, Dean DC. Liu Y, et al. Invest Ophthalmol Vis Sci. 2009 Nov;50(11):5080-8. doi: 10.1167/iovs.08-2911. Epub 2009 Jun 10. Invest Ophthalmol Vis Sci. 2009. PMID: 19515996 Free PMC article.
References
- Bear, J., Y. Hong, and M. Schartl. 2003. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 130:6545–6553. - PubMed
- Chin, L., G. Merlino, and R.A. DePinho. 1998. Malignant melanoma: modern black plague and genetic black box. Genes Dev. 12:3467–3481. - PubMed
- Harbour, J.W., L. Worley, D. Ma, and M. Cohen. 2002. Transducible peptide therapy for uveal melanoma and retinoblastoma. Arch. Ophthalmol. 120:1341–1346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources