The net of life: reconstructing the microbial phylogenetic network - PubMed (original) (raw)
Comparative Study
The net of life: reconstructing the microbial phylogenetic network
Victor Kunin et al. Genome Res. 2005 Jul.
Abstract
It has previously been suggested that the phylogeny of microbial species might be better described as a network containing vertical and horizontal gene transfer (HGT) events. Yet, all phylogenetic reconstructions so far have presented microbial trees rather than networks. Here, we present a first attempt to reconstruct such an evolutionary network, which we term the "net of life". We use available tree reconstruction methods to infer vertical inheritance, and use an ancestral state inference algorithm to map HGT events on the tree. We also describe a weighting scheme used to estimate the number of genes exchanged between pairs of organisms. We demonstrate that vertical inheritance constitutes the bulk of gene transfer on the tree of life. We term the bulk of horizontal gene flow between tree nodes as "vines", and demonstrate that multiple but mostly tiny vines interconnect the tree. Our results strongly suggest that the HGT network is a scale-free graph, a finding with important implications for genome evolution. We propose that genes might propagate extremely rapidly across microbial species through the HGT network, using certain organisms as hubs.
Figures
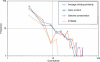
Figure 1.
Distribution of HGT vine widths.
Figure 2.
Connectivity of the HGT network.
Figure 3.
Three-dimensional representation of the net of life. The tree backbone was generated by using the average gene similarity approach (see Methods). The root is represented as a yellow sphere. Bacteria are shown as nodes on cyan branches; Archaea, as nodes on green branches. Red lines correspond to the vines representing HGT. The radius of the nodes is proportional to the estimated gene content size (in terms of number of gene families). Also, the widths of both the vertical inheritance branches and the horizontal inheritance vines correspond to the numbers of gene families transferred by either mechanism. For visualization purposes, only values for HGT vine width >30 are shown. Certain key species and taxa are labeled; for full names, please refer to Supplemental material.
Similar articles
- Inferring phylogenetic networks by the maximum parsimony criterion: a case study.
Jin G, Nakhleh L, Snir S, Tuller T. Jin G, et al. Mol Biol Evol. 2007 Jan;24(1):324-37. doi: 10.1093/molbev/msl163. Epub 2006 Oct 26. Mol Biol Evol. 2007. PMID: 17068107 - A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm.
Podell S, Gaasterland T, Allen EE. Podell S, et al. BMC Bioinformatics. 2008 Oct 7;9:419. doi: 10.1186/1471-2105-9-419. BMC Bioinformatics. 2008. PMID: 18840280 Free PMC article. - A model of horizontal gene transfer and the bacterial phylogeny problem.
Galtier N. Galtier N. Syst Biol. 2007 Aug;56(4):633-42. doi: 10.1080/10635150701546231. Syst Biol. 2007. PMID: 17661231 - Phylogenomic networks.
Dagan T. Dagan T. Trends Microbiol. 2011 Oct;19(10):483-91. doi: 10.1016/j.tim.2011.07.001. Epub 2011 Aug 3. Trends Microbiol. 2011. PMID: 21820313 Review. - Detecting lateral genetic transfer : a phylogenetic approach.
Beiko RG, Ragan MA. Beiko RG, et al. Methods Mol Biol. 2008;452:457-69. doi: 10.1007/978-1-60327-159-2_21. Methods Mol Biol. 2008. PMID: 18566777 Review.
Cited by
- Adapting the rhizome concept to an extended definition of viral quasispecies and the implications for molecular evolution.
Landa CR, Ariza-Mateos A, Briones C, Perales C, Wagner A, Domingo E, Gómez J. Landa CR, et al. Sci Rep. 2024 Aug 2;14(1):17914. doi: 10.1038/s41598-024-68760-6. Sci Rep. 2024. PMID: 39095425 Free PMC article. - CGG toolkit: Software components for computational genomics.
Vasileiou D, Karapiperis C, Baltsavia I, Chasapi A, Ahrén D, Janssen PJ, Iliopoulos I, Promponas VJ, Enright AJ, Ouzounis CA. Vasileiou D, et al. PLoS Comput Biol. 2023 Nov 7;19(11):e1011498. doi: 10.1371/journal.pcbi.1011498. eCollection 2023 Nov. PLoS Comput Biol. 2023. PMID: 37934729 Free PMC article. - Pan-genomic comparison of a potential solvent-tolerant alkaline protease-producing Exiguobacterium sp. TBG-PICH-001 isolated from a marine habitat.
Srivastava N, Shiburaj S, Khare SK. Srivastava N, et al. 3 Biotech. 2023 Nov;13(11):371. doi: 10.1007/s13205-023-03796-5. Epub 2023 Oct 16. 3 Biotech. 2023. PMID: 37854939 Free PMC article. - Dissecting the HGT network of carbon metabolic genes in soil-borne microbiota.
Li L, Liu Y, Xiao Q, Xiao Z, Meng D, Yang Z, Deng W, Yin H, Liu Z. Li L, et al. Front Microbiol. 2023 Jul 7;14:1173748. doi: 10.3389/fmicb.2023.1173748. eCollection 2023. Front Microbiol. 2023. PMID: 37485539 Free PMC article.
References
- Alfano, J.R. and Collmer, A. 2004. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42: 385-414. - PubMed
- Bapteste, E., Boucher, Y., Leigh, J., and Doolittle, W.F. 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12: 406-411. - PubMed
- Boucher, Y., Douady, C.J., Papke, R.T., Walsh, D.A., Boudreau, M.E., Nesbo, C.L., Case, R.J., and Doolittle, W.F. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37: 283-328. - PubMed
- Brown, J.R., Douady, C.J., Italia, M.J., Marshall, W.E., and Stanhope, M.J. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28: 281-285. - PubMed
Web site references
- http://cgg.ebi.ac.uk/services/ortho-fam/; OFAM data set.
- http://cgg.ebi.ac.uk/cgi-bin/gps/GPS.pl; Genome Phylogeny Server.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources